Centromere
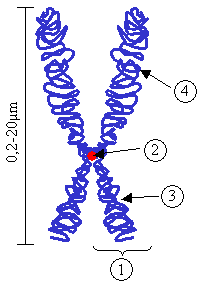
(2) Centromere. The point where the two chromatids touch.
(3) Short arm.
(4) Long arm.
Because the arms are unequal, this chromosome is called "submetacentric."
The centromere is a region, often found in the middle of the chromosome, involved in cell division and the control of gene expression.
Function
The centromere is, together with telomeres and origin of replications, one of the essential parts of any eukaryotic chromosome. The centromere usually contains specific types of DNA sequences which are in higher eukaryotes typically tandem repetitive sequences, often called "satellite DNA". These sequences bind specific proteins called "cen"-Proteins. During mitosis the centromeres can be identified in particular during the metaphase stage as a constriction at the chromosome. At this centromeric constriction the two mostly identical halves of the chromosome, the sister chromatids, are held together until late metaphase. During mitotic division, a transient structure called kinetochore is formed on top of the centromeres. The kinetochores are the sites where the spindle fibers attach. Kinetochores and the spindle apparatus are responsible for the movement of the two sister chromatids to opposite poles of dividing cell nucleus during anaphase. Usually the mitosis is immediately followed by a cell division cytokinesis. However, mitosis and cytokinesis are separate processes and can be uncoupled.
A centromere functions in sister chromatid adhesion, kinetochore formation, and pairing of homologous chromosomes during mitosis, prophase and metaphase. The centromere is also where kinetochore formation takes place: proteins bind on the centromeres that form an anchor point for the spindle formation required for the pull of chromosomes toward the centrioles during anaphase and telophase of mitosis.
Improperly functioning centromeres result in the chromosomes that do not align and separate properly, resulting in aneuploidy or daughter cells receiving the wrong number of chromosomes. Aneuploidy can cause conditions such as Down syndrome if the cells survive at all. [1]
Centromere Positions
Each chromosome has two arms, labeled p (the shorter of the two) and q (the longer). They can be connected in either metacentric, submetacentric, acrocentric or telocentric manner. (While the p arm is named for petite meaning small, the q arm is named thus simply because it follows p in the alphabet.)
Metacentric
If both arms are equal in length, the chromosome is said to be metacentric. Robertsonian -- fusion of 2 p arms by centric fusion to form metacentric chromosome.
Submetacentric
If arms' lengths are unequal, the chromosome is said to be submetacentric
Acrocentric
If the p (short) arm is so short that is hard to observe, but still present, then the chromosome is acrocentric (The "acro-" in acrocentric refers to the Greek word for "peak.").
There are five acrocentric chromosomes in the human genome: 13, 14, 15, 21 and 22. These five chromosomes are the site of genes encoding rRNAs.
Telocentric
A telocentric chromosome's centromere is located at the terminal end of the chromosome. Telomeres may extend from both ends of the chromosome. All mouse chromosomes are telocentric [2]; in humans, the Y sex chromosome is telocentric.
Holocentric
With holocentric chromosomes, the entire length of the chromosome acts as the centromere. Examples of this type of centromere can be found scattered throughout the plant and animal kingdoms[3] with the most well known example being in the worm, Caenorhabditis elegans.
The centromeric sequence
There are two types of centromeres[4]. In regional centromeres, DNA sequences contribute to but do not define function. Regional centromeres contain large amounts of DNA and are often packaged into heterochromatin. In most eukaryotes, the centromere has no defined DNA sequence. It typically consists of large arrays of repetitive DNA (e.g. satellite DNA) where the sequence within individual repeat elements is similar but not identical. In humans, the primary centromeric repeat unit is called α-satellite (or alphoid), although a number of other sequence types are found in this region.
Point centromeres are smaller and more compact. DNA sequences are both necessary and sufficient to specify centromere identity and function in organisms with point centromeres. In budding yeasts, the centromere region is relatively small (about 125 bp DNA) and contains two highly conserved DNA sequences that serve as binding sites for essential kinetochore proteins.
Inheritance
Epigenetic inheritance plays a major role in specifying the centromere in most organisms. The daughter chromosomes will assemble centromeres in the same place as the parent chromosome, independent of sequence. However, there must still be some original way in which the centromere is specified, even if it is subsequently propagated epigenetically.
Structure
The centromeric DNA is normally in a heterochromatin state, which is essential for the recruitment of the cohesin complex that mediates sister chromatid cohesion after DNA replication as well as coordinating sister chromatid separation during anaphase. In this chromatin, the normal histone H3 is replaced with a centromere-specific variant, CENP-A in humans (Lodish et al. 2004). The presence of CENP-A is believed to be important for the assembly of the kinetochore on the centromere. CENP-C has been shown to localise almost exclusively to these regions of CENP-A associated chromatin.
In the yeast Schizosaccharomyces pombe (and probably in other eukaryotes), the formation of centromeric heterochromatin is connected to RNAi.[5] In nematodes such as Caenorhabditis elegans, some plants, and the insect orders Lepidoptera and Hemiptera, chromosomes are "holocentric", indicating that there is not a primary site of microtubule attachments or a primary constriction, and a "diffuse" kinetochore assembles along the entire length of the chromosome.
Centromeric aberrations
In rare cases in humans, neocentromeres can form at new sites on the chromosome. This must be coupled with the inactivation of the previous centromere since chromosomes with two functional centromeres (Dicentric chromosome) will result in chromosome breakage during mitosis. In some unusual cases human neocentromeres have been observed to form spontaneously on fragmented chromosomes. Some of these new positions were originally euchromatic and lack alpha satellite DNA altogether.
Centromere proteins are also the autoantigenic target for some anti-nuclear antibodies such as anti-centromere antibodies
Related links
References
- ^ Earnshaw et al., 1989 W.C. Earnshaw, H. Ratrie 3rd and G. Stetten, Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads, Chromosoma 98 (1989), pp. 1–12
- ^ Mouse Genome Sequencing and Comparative Analysis of the Mouse Genome, Nature, 2002
- ^ Abby F. Dernburg. (2001). Here, There, and Everywhere Kinetochore Function on Holocentric Chromosomes. J Cell Biol. 153(6): f33–f38. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=11402076
- ^ Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. (1995). The centromere: hub of chromosomal activities. Science. 270(5242):1591-4. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=Pubmed&term=7502067
- ^ Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002-09-13). "Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi". Science: 297(5588):1818–9. doi:10.1126/science.1074973.
{{cite journal}}: Check|doi=value (help); Check date values in:|date=(help); Unknown parameter|doi_brokendate=ignored (|doi-broken-date=suggested) (help)CS1 maint: multiple names: authors list (link)
Further reading
- Lodish et al.; Molecular Cell Biology; fifth edition; 2004; ISBN 0716743663
