Histone: Difference between revisions
| Line 42: | Line 42: | ||
The core histones all exist as [[Protein dimer|dimer]]s, which are similar in that they all possess the histone fold domain; three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif).<ref>{{cite journal | vauthors = Mariño-Ramírez L, Kann MG, Shoemaker BA, Landsman D | title = Histone structure and nucleosome stability | journal = Expert Review of Proteomics | volume = 2 | issue = 5 | pages = 719–29 | date = Oct 2005 | pmid = 16209651 | pmc = 1831843 | doi = 10.1586/14789450.2.5.719 }}</ref> The resulting four distinct dimers then come together to form one octameric [[nucleosome]] core, approximately 63 Angstroms in diameter (a [[solenoid (DNA)]]-like particle). Around 146 [[base pairs]] (bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across.<ref name=pmid9305837>{{cite journal | vauthors = Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ | title = Crystal structure of the nucleosome core particle at 2.8 A resolution | journal = Nature | volume = 389 | issue = 6648 | pages = 251–60 | date = Sep 1997 | pmid = 9305837 | doi = 10.1038/38444 }} {{PDB|1AOI}}</ref> The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place<ref name="isbn0-915274-84-1">{{cite book | last = Farkas | first = Daniel |title=DNA simplified: the hitchhiker's guide to DNA |publisher=AACC Press |location=Washington, D.C |year=1996 |isbn=0-915274-84-1 | name-list-format = vanc }}</ref> and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of [[DNA]] separating each pair of [[nucleosome]]s (also referred to as linker [[DNA]]). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed [[chromosome]]s are assembled through interactions between nucleosomes and other regulatory proteins. |
The core histones all exist as [[Protein dimer|dimer]]s, which are similar in that they all possess the histone fold domain; three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif).<ref>{{cite journal | vauthors = Mariño-Ramírez L, Kann MG, Shoemaker BA, Landsman D | title = Histone structure and nucleosome stability | journal = Expert Review of Proteomics | volume = 2 | issue = 5 | pages = 719–29 | date = Oct 2005 | pmid = 16209651 | pmc = 1831843 | doi = 10.1586/14789450.2.5.719 }}</ref> The resulting four distinct dimers then come together to form one octameric [[nucleosome]] core, approximately 63 Angstroms in diameter (a [[solenoid (DNA)]]-like particle). Around 146 [[base pairs]] (bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across.<ref name=pmid9305837>{{cite journal | vauthors = Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ | title = Crystal structure of the nucleosome core particle at 2.8 A resolution | journal = Nature | volume = 389 | issue = 6648 | pages = 251–60 | date = Sep 1997 | pmid = 9305837 | doi = 10.1038/38444 }} {{PDB|1AOI}}</ref> The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place<ref name="isbn0-915274-84-1">{{cite book | last = Farkas | first = Daniel |title=DNA simplified: the hitchhiker's guide to DNA |publisher=AACC Press |location=Washington, D.C |year=1996 |isbn=0-915274-84-1 | name-list-format = vanc }}</ref> and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of [[DNA]] separating each pair of [[nucleosome]]s (also referred to as linker [[DNA]]). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed [[chromosome]]s are assembled through interactions between nucleosomes and other regulatory proteins. |
||
Histones are subdivided into canonical replication-dependent histones that are expressed during the S-phase of cell cycle and replication-independent [[histone variants]], expressed during the whole cell cycle. In animals, genes encoding canonical histones are typically clustered along the chromosome, lack introns and use a stem loop structure at the 3’ end instead of a polyA tail. Genes encoding histone variants are usually not clustered, have introns and their mRNAs are regulated with polyA tails. Complex multicellular organisms typically have a higher number of histone variants providing a variety of different functions. Recent data are accumulating about the roles of diverse histone variants highlighting the functional links between variants and the delicate regulation of organism development. Histone variants from different organisms, their classification and variant specific features can be found in [https://www.ncbi.nlm.nih.gov/projects/HistoneDB2.0/ "HistoneDB 2.0 - Variants"] database. |
Histones are subdivided into canonical replication-dependent histones that are expressed during the S-phase of cell cycle and replication-independent [[histone variants]], expressed during the whole cell cycle. In animals, genes encoding canonical histones are typically clustered along the chromosome, lack introns and use a stem loop structure at the 3’ end instead of a polyA tail. Genes encoding histone variants are usually not clustered, have introns and their mRNAs are regulated with polyA tails. Complex multicellular organisms typically have a higher number of histone variants providing a variety of different functions. Recent data are accumulating about the roles of diverse [[histone variants]] highlighting the functional links between variants and the delicate regulation of organism development. [[Histone variants]] from different organisms, their classification and variant specific features can be found in [https://www.ncbi.nlm.nih.gov/projects/HistoneDB2.0/ "HistoneDB 2.0 - Variants"] database. |
||
The following is a list of human histone proteins: |
The following is a list of human histone proteins: |
||
Revision as of 20:29, 1 February 2017
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes.[1][2] They are the chief protein components of chromatin, acting as spools around which DNA winds, and playing a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to 1 in human DNA). For example, each human diploid cell (containing 23 pairs of chromosomes) has about 1.8 meters of DNA, but wound on the histones it has about 90 micrometers (0.09 mm) of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes.[3]
| Core histone H2A/H2B/H3/H4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
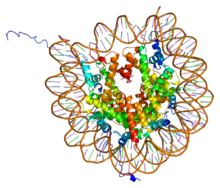 PDB rendering of Complex between nucleosome core particle (h3,h4,h2a,h2b) and 146 bp long DNA fragment based on 1aoi. | |||||||||||
| Identifiers | |||||||||||
| Symbol | Histone | ||||||||||
| Pfam | PF00125 | ||||||||||
| Pfam clan | CL0012 | ||||||||||
| InterPro | IPR007125 | ||||||||||
| SCOP2 | 1hio / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| linker histone H1 and H5 family | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Linker_histone | ||||||||||
| Pfam | PF00538 | ||||||||||
| InterPro | IPR005818 | ||||||||||
| SMART | SM00526 | ||||||||||
| SCOP2 | 1hst / SCOPe / SUPFAM | ||||||||||
| |||||||||||
Classes and histone variants
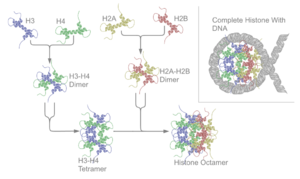
Five major families of histones exist: H1/H5, H2A, H2B, H3, and H4.[2][4][5][6] Histones H2A, H2B, H3 and H4 are known as the core histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain; three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif).[7] The resulting four distinct dimers then come together to form one octameric nucleosome core, approximately 63 Angstroms in diameter (a solenoid (DNA)-like particle). Around 146 base pairs (bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across.[8] The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place[9] and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA separating each pair of nucleosomes (also referred to as linker DNA). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
Histones are subdivided into canonical replication-dependent histones that are expressed during the S-phase of cell cycle and replication-independent histone variants, expressed during the whole cell cycle. In animals, genes encoding canonical histones are typically clustered along the chromosome, lack introns and use a stem loop structure at the 3’ end instead of a polyA tail. Genes encoding histone variants are usually not clustered, have introns and their mRNAs are regulated with polyA tails. Complex multicellular organisms typically have a higher number of histone variants providing a variety of different functions. Recent data are accumulating about the roles of diverse histone variants highlighting the functional links between variants and the delicate regulation of organism development. Histone variants from different organisms, their classification and variant specific features can be found in "HistoneDB 2.0 - Variants" database.
The following is a list of human histone proteins:
| Super family | Family | Subfamily | Members |
|---|---|---|---|
| Linker | H1 | H1F | H1F0, H1FNT, H1FOO, H1FX |
| H1H1 | HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H1T | ||
| Core | H2A | H2AF | H2AFB1, H2AFB2, H2AFB3, H2AFJ, H2AFV, H2AFX, H2AFY, H2AFY2, H2AFZ |
| H2A1 | HIST1H2AA, HIST1H2AB, HIST1H2AC, HIST1H2AD, HIST1H2AE, HIST1H2AG, HIST1H2AI, HIST1H2AJ, HIST1H2AK, HIST1H2AL, HIST1H2AM | ||
| H2A2 | HIST2H2AA3, HIST2H2AC | ||
| H2B | H2BF | H2BFM, H2BFS, H2BFWT | |
| H2B1 | HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO | ||
| H2B2 | HIST2H2BE | ||
| H3 | H3A1 | HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J | |
| H3A2 | HIST2H3C | ||
| H3A3 | HIST3H3 | ||
| H4 | H41 | HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L | |
| H44 | HIST4H4 |
Structure
The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer, forming two nearly symmetrical halves by tertiary structure (C2 symmetry; one macromolecule is the mirror image of the other).[8] The H2A-H2B dimers and H3-H4 tetramer also show pseudodyad symmetry. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution, all featuring a 'helix turn helix turn helix' motif (which allows the easy dimerisation). They also share the feature of long 'tails' on one end of the amino acid structure - this being the location of post-translational modification (see below).
It has been proposed that histone proteins are evolutionarily related to the helical part of the extended AAA+ ATPase domain, the C-domain, and to the N-terminal substrate recognition domain of Clp/Hsp100 proteins. Despite the differences in their topology, these three folds share a homologous helix-strand-helix (HSH) motif.[10]
Using an electron paramagnetic resonance spin-labeling technique, British researchers measured the distances between the spools around which eukaryotic cells wind their DNA. They determined the spacings range from 59 to 70 Å.[11]
In all, histones make five types of interactions with DNA:
- Helix-dipoles form alpha-helixes in H2B, H3, and H4 cause a net positive charge to accumulate at the point of interaction with negatively charged phosphate groups on DNA
- Hydrogen bonds between the DNA backbone and the amide group on the main chain of histone proteins
- Nonpolar interactions between the histone and deoxyribose sugars on DNA
- Salt bridges and hydrogen bonds between side chains of basic amino acids (especially lysine and arginine) and phosphate oxygens on DNA
- Non-specific minor groove insertions of the H3 and H2B N-terminal tails into two minor grooves each on the DNA molecule
The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to their water solubility.
Histones are subject to post translational modification by enzymes primarily on their N-terminal tails, but also in their globular domains.[12][13] Such modifications include methylation, citrullination, acetylation, phosphorylation, SUMOylation, ubiquitination, and ADP-ribosylation. This affects their function of gene regulation.
In general, genes that are active have less bound histone, while inactive genes are highly associated with histones during interphase[citation needed]. It also appears that the structure of histones has been evolutionarily conserved, as any deleterious mutations would be severely maladaptive. All histones have a highly positively charged N-terminus with many lysine and arginine residues.
History
Histones were discovered in 1884 by Albrecht Kossel. The word "histone" dates from the late 19th century and is from the German word "Histon", a word itself of uncertain origin - perhaps from the Greek histanai or histos. Until the early 1990s, histones were dismissed by most as inert packing material for eukaryotic nuclear DNA, a view based in part on the models of Mark Ptashne and others, who believed that transcription was activated by protein-DNA and protein-protein interactions on largely naked DNA templates, as is the case in bacteria.
During the 1980s, Yahli Lorch and Roger Kornberg[14] showed that a nucleosome on a core promoter prevents the initiation of transcription in vitro, and Michael Grunstein[15] demonstrated that histones repress transcription in vivo, leading to the idea of the nucleosome as a general gene repressor. Relief from repression is believed to involve both histone modification and the action of chromatin-remodeling complexes. Vincent Allfrey and Alfred Mirsky earlier proposed a role of histone modification in transcriptional activation,[16] regarded as a molecular manifestation of epigenetics. Michael Grunstein[17] and David Allis[18] found support for this proposal, in the importance of histone acetylation for transcription in yeast and the activity of the transcriptional activator Gcn5 as a histone acetyltransferase.
The discovery of the H5 histone appears to date back to the 1970s,[19] and it is now considered an isoform of Histone H1.[2][4][5][6]
Conservation across species
Histones are found in the nuclei of eukaryotic cells, and in certain Archaea, namely Thermoproteales and Euryarchaea, but not in bacteria. The unicellular algae known as dinoflagellates were previously thought to be the only eukaryotes that completely lack histones,[20] however, later studies showed that their DNA still encodes histone genes.[21]
Archaeal histones may well resemble the evolutionary precursors to eukaryotic histones. Histone proteins are among the most highly conserved proteins in eukaryotes, emphasizing their important role in the biology of the nucleus.[2]: 939 In contrast mature sperm cells largely use protamines to package their genomic DNA, most likely because this allows them to achieve an even higher packaging ratio.[22]
Core histones are highly conserved proteins; that is, there are very few differences among the amino acid sequences of the histone proteins of different species.
There are some variant forms in some of the major classes. They share amino acid sequence homology and core structural similarity to a specific class of major histones but also have their own feature that is distinct from the major histones. These minor histones usually carry out specific functions of the chromatin metabolism. For example, histone H3-like CenpA is associated with only the centromere region of the chromosome. Histone H2A variant H2A.Z is associated with the promoters of actively transcribed genes and also involved in the prevention of the spread of silent heterochromatin.[23] Furthermore, H2A.Z has roles in chromatin for genome stability.[24] Another H2A variant H2A.X is phosphorylated at S139 in regions around double-strand breaks and marks the region undergoing DNA repair.[25] Histone H3.3 is associated with the body of actively transcribed genes.[26]
Function
Compacting DNA strands
Histones act as spools around which DNA winds. This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nuclei: the compacted molecule is 40,000 times shorter than an unpacked molecule.
Chromatin regulation
Histones undergo posttranslational modifications that alter their interaction with DNA and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome, which can be covalently modified at several places. Modifications of the tail include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, citrullination, and ADP-ribosylation. The core of the histones H2A and H2B can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code".[27][28] Histone modifications act in diverse biological processes such as gene regulation, DNA repair, chromosome condensation (mitosis) and spermatogenesis (meiosis).[29]
The common nomenclature of histone modifications is:
- The name of the histone (e.g., H3)
- The single-letter amino acid abbreviation (e.g., K for Lysine) and the amino acid position in the protein
- The type of modification (Me: methyl, P: phosphate, Ac: acetyl, Ub: ubiquitin)
- The number of modifications (only Me is known to occur in more than one copy per residue. 1, 2 or 3 is mono-, di- or tri-methylation)
So H3K4me1 denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal) of the H3 protein.
Examples of histone modifications in transcription regulation include:
| Type of modification |
Histone | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| H3K4 | H3K9 | H3K14 | H3K27 | H3K79 | H3K36 | H4K20 | H2BK5 | H2BK20 | |
| mono-methylation | activation[30] | activation[31] | activation[31] | activation[31][32] | activation[31] | activation[31] | |||
| di-methylation | repression[33] | repression[33] | activation[32] | ||||||
| tri-methylation | activation[34] | repression[31] | repression[31] | activation,[32] repression[31] |
activation | repression[33] | |||
| acetylation | activation[34] | activation[34] | activation[35] | activation | |||||
Functions of histone modifications

A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a histone code, whereby combinations of histone modifications have specific meanings. However, most functional data concerns individual prominent histone modifications that are biochemically amenable to detailed study.
Chemistry of histone modifications
Lysine methylation
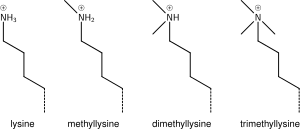
The addition of one, two or three methyl groups to lysine has little effect on the chemistry of the histone; methylation leaves the charge of the lysine intact and adds a minimal number of atoms so steric interactions are mostly unaffected. However, proteins containing Tudor, chromo or PHD domains, amongst others, can recognise lysine methylation with exquisite sensitivity and differentiate mono, di and tri-methyl lysine, to the extent that, for some lysines (e.g.: H4K20) mono, di and tri-methylation appear to have different meanings. Because of this, lysine methylation tends to be a very informative mark and dominates the known histone modification functions.
Arginine methylation
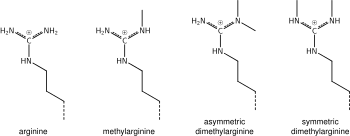
What was said above of the chemistry of lysine methylation also applies to arginine methylation, and some protein domains—e.g., Tudor domains—can be specific for methyl arginine instead of methyl lysine. Arginine is known to be mono- or di-methylated, and methylation can be symmetric or asymmetric, potentially with different meanings.
Arginine citrullination
Enzymes called peptidylarginine deiminases (PADs) hydrolyze the imine group of arginines and attach a keto group, so that there is one less positive charge on the amino acid residue. This process has been involved in the activation of gene expression by making the modified histones less tightly bound to DNA and thus making the chromatin more accessible.[36] PADs can also produce the opposite effect by removing or inhibiting mono-methylation of arginine residues on histones and thus antagonizing the positive effect arginine methylation has on transcriptional activity.[37]
Lysine acetylation

Addition of an acetyl group has a major chemical effect on lysine as it neutralises the positive charge. This reduces electrostatic attraction between the histone and the negatively charged DNA backbone, loosening the chromatin structure; highly acetylated histones form more accessible chromatin and tend to be associated with active transcription. Lysine acetylation appears to be less precise in meaning than methylation, in that histone acetyltransferases tend to act on more than one lysine; presumably this reflects the need to alter multiple lysines to have a significant effect on chromatin structure. The modification includes H3K27ac.
Serine/threonine/tyrosine phosphorylation
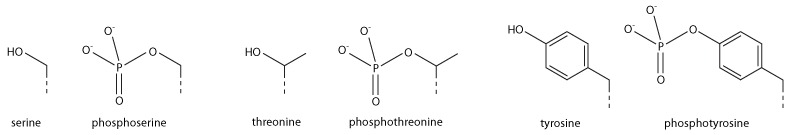
Addition of a negatively charged phosphate group can lead to major changes in protein structure, leading to the well-characterised role of phosphorylation in controlling protein function. It is not clear what structural implications histone phosphorylation has, but histone phosphorylation has clear functions as a post-translational modification, and binding domains such as BRCT have been characterised.
Functions in transcription
Most well-studied histone modifications are involved in control of transcription.
Actively transcribed genes
Two histone modifications are particularly associated with active transcription:
- Trimethylation of H3 lysine 4 (H3K4Me3)
- This trimethylation occurs at the promoter of active genes[38][39][40] and is performed by the COMPASS complex.[41][42][43] Despite the conservation of this complex and histone modification from yeast to mammals, it is not entirely clear what role this modification plays. However, it is an excellent mark of active promoters and the level of this histone modification at a gene’s promoter is broadly correlated with transcriptional activity of the gene. The formation of this mark is tied to transcription in a rather convoluted manner: early in transcription of a gene, RNA polymerase II undergoes a switch from initiating’ to ‘elongating’, marked by a change in the phosphorylation states of the RNA polymerase II C terminal domain (CTD). The same enzyme that phosphorylates the CTD also phosphorylates the Rad6 complex,[44][45] which in turn adds a ubiquitin mark to H2B K123 (K120 in mammals).[46] H2BK123Ub occurs throughout transcribed regions, but this mark is required for COMPASS to trimethylate H3K4 at promoters.[47][48]
- Trimethylation of H3 lysine 36 (H3K36Me3)
- This trimethylation occurs in the body of active genes and is deposited by the methyltransferase Set2.[49] This protein associates with elongating RNA polymerase II, and H3K36Me3 is indicative of actively transcribed genes.[50] H3K36Me3 is recognised by the Rpd3 histone deacetylase complex, which removes acetyl modifications from surrounding histones, increasing chromatin compaction and repressing spurious transcription.[51][52][53] Increased chromatin compaction prevents transcription factors from accessing DNA, and reduces the likelihood of new transcription events being initiated within the body of the gene. This process therefore helps ensure that transcription is not interrupted.
Repressed genes
Three histone modifications are particularly associated with repressed genes:
- Trimethylation of H3 lysine 27 (H3K27me3)
- This histone modification is depositied by the polycomb complex PRC2.[54] It is a clear marker of gene repression,[55] and is likely bound by other proteins to exert a repressive function. Another polycomb complex, PRC1, can bind H3K27me3[55] and adds the histone modification H2AK119Ub which aids chromatin compaction.[56][57] Based on this data it appears that PRC1 is recruited through the action of PRC2, however, recent studies show that PRC1 is recruited to the same sites in the absence of PRC2.[58][59]
- Di and tri-methylation of H3 lysine 9 (H3K9me2/3)
- H3K9me2/3 is a well-characterised marker for heterochromatin, and is therefore strongly associated with gene repression. The formation of heterochromatin has been best studied in the yeast Schizosaccharomyces pombe, where it is initiated by recruitment of the RNA-induced transcriptional silencing complex to double stranded RNAs produced from centromeric repeats.[60] RITS recruits the Clr4 histone methyltransferase which deposits H3K9me2/3.[61] This process is called histone methylation. H3K9Me2/3 serves as a binding site for the recruitment of Swi6 (heterochromatin protein 1 or HP1, another classic heterochromatin marker)[62][63] which in turn recruits further repressive activities including histone modifiers such as histone deacetylases and histone methyltransferases.
- Trimethylation of H4 lysine 20 (H4K20me3)
- This modification is tightly associated with heterochromatin,[64][65] although its functional importance remains unclear. This mark is placed by the Suv4-20h methyltransferase, which is at least in part recruited by heterochromatin protein 1.[64]
Bivalent promoters
Analysis of histone modifications in embryonic stem cells (and other stem cells) revealed many gene promoters carrying both H3K4Me3 and H3K27Me3, in other words these promoters display both activating and repressing marks simultaneously. This peculiar combination of modifications marks genes that are poised for transcription; they are not required in stem cells, but are rapidly required after differentiation into some lineages. Once the cell starts to differentiate, these bivalent promoters are resolved to either active or repressive states depending on the chosen lineage.[66]
Other functions
DNA damage
Marking sites of DNA damage is an important function for histone modifications. It also protects DNA from getting destroyed by ultraviolet radiation of sun.
- Phosphorylation of H2AX at serine 139 (γH2AX)
- Phosphorylated H2AX (also known as gamma H2AX) is a marker for DNA double strand breaks,[67] and forms part of the response to DNA damage.[25][68] H2AX is phosphorylated early after detection of DNA double strand break, and forms a domain extending many kilobases either side of the damage.[67][69][70] Gamma H2AX acts as a binding site for the protein MDC1, which in turn recruits key DNA repair proteins[71] (this complex topic is well reviewed in[72]) and as such, gamma H2AX forms a vital part of the machinery that ensures genome stability.
- Acetylation of H3 lysine 56 (H3K56Ac)
- H3K56Acx is required for genome stability.[73][74] H3K56 is acetylated by the p300/Rtt109 complex,[75][76][77] but is rapidly deacetylated around sites of DNA damage. H3K56 acetylation is also required to stabilise stalled replication forks, preventing dangerous replication fork collapses.[78][79] Although in general mammals make far greater use of histone modifications than microorganisms, a major role of H3K56Ac in DNA replication exists only in fungi, and this has become a target for antibiotic development.[80]
Chromosome condensation
- Phosphorylation of H3 at serine 10 (phospho-H3S10)
- The mitotic kinase aurora B phosphorylates histone H3 at serine 10, triggering a cascade of changes that mediate mitotic chromosome condensation.[81][82] Condensed chromosomes therefore stain very strongly for this mark, but H3S10 phosphorylation is also present at certain chromosome sites outside mitosis, for example in pericentric heterochromatin of cells during G2. H3S10 phosphorylation has also been linked to DNA damage caused by R loop formation at highly transcribed sites.[83]
- Phosphorylation H2B at serine 10/14 (phospho-H2BS10/14)
- Phosphorylation of H2B at serine 10 (yeast) or serine 14 (mammals) is also linked to chromatin condensation, but for the very different purpose of mediating chromosome condensation during apoptosis.[84][85] This mark is not simply a late acting bystander in apoptosis as yeast carrying mutations of this residue are resistant to hydrogen peroxide-induced apoptotic cell death.
See also
References
- ^ Youngson, Robert M. (2006). Collins Dictionary of Human Biology. Glasgow: HarperCollins. ISBN 0-00-722134-7.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ a b c d Cox, Michael; Nelson, David R.; Lehninger, Albert L (2005). Lehninger Principles of Biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-4339-6.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (Apr 2002). "Histone H2A variants H2AX and H2AZ". Current Opinion in Genetics & Development. 12 (2): 162–9. doi:10.1016/S0959-437X(02)00282-4. PMID 11893489.
- ^ a b "Histone Variants Database 2.0". National Center for Biotechnology Information. Retrieved 13 January 2017.
- ^ a b Bhasin M, Reinherz EL, Reche PA (2006). "Recognition and classification of histones using support vector machine". Journal of Computational Biology. 13 (1): 102–12. doi:10.1089/cmb.2006.13.102. PMID 16472024.
- ^ a b Hartl, Daniel L.; Freifelder, David; Snyder, Leon A. (1988). Basic Genetics. Boston: Jones and Bartlett Publishers. ISBN 0-86720-090-1.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Mariño-Ramírez L, Kann MG, Shoemaker BA, Landsman D (Oct 2005). "Histone structure and nucleosome stability". Expert Review of Proteomics. 2 (5): 719–29. doi:10.1586/14789450.2.5.719. PMC 1831843. PMID 16209651.
- ^ a b Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (Sep 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature. 389 (6648): 251–60. doi:10.1038/38444. PMID 9305837. PDB: 1AOI
- ^ Farkas, Daniel (1996). DNA simplified: the hitchhiker's guide to DNA. Washington, D.C: AACC Press. ISBN 0-915274-84-1.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Alva V, Ammelburg M, Söding J, Lupas AN (March 2007). "On the origin of the histone fold". BMC Structural Biology. 7: 17. doi:10.1186/1472-6807-7-17. PMC 1847821. PMID 17391511.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
- ^ Ward R, Bowman A, El-Mkami H, Owen-Hughes T, Norman DG (Feb 2009). "Long distance PELDOR measurements on the histone core particle". Journal of the American Chemical Society. 131 (4): 1348–9. doi:10.1021/ja807918f. PMC 3501648. PMID 19138067.
- ^ Mersfelder EL, Parthun MR (19 May 2006). "The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure". Nucleic Acids Research. 34 (9): 2653–62. doi:10.1093/nar/gkl338. PMC 1464108. PMID 16714444.
- ^ Tropberger P, Schneider R (Jun 2013). "Scratching the (lateral) surface of chromatin regulation by histone modifications". Nature Structural & Molecular Biology. 20 (6): 657–61. doi:10.1038/nsmb.2581. PMID 23739170.
- ^ Lorch Y, LaPointe JW, Kornberg RD (Apr 1987). "Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones". Cell. 49 (2): 203–10. doi:10.1016/0092-8674(87)90561-7. PMID 3568125.
- ^ Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M (Oct 1988). "Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast". Cell. 55 (1): 27–39. doi:10.1016/0092-8674(88)90006-2. PMID 3048701.
- ^ Allfrey, Vincent (1966). "RNA synthesis and histone acetylation during the course of gene activation in lymphocytes". Proc Natl Acad Sci U S A. 55: 805–812. doi:10.1073/pnas.55.4.805.
- ^ Grunstein, Michael (1991). "Yeast histone H4 N-terminal sequence is required for promoter activation in vivo". Cell. 65: 1023–1031. doi:10.1016/0092-8674(91)90554-c.
- ^ Allis, C David (1996). "Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation". Cell. 84: 843–851. doi:10.1016/s0092-8674(00)81063-6.
- ^ Aviles FJ, Chapman GE, Kneale GG, Crane-Robinson C, Bradbury EM (Aug 1978). "The conformation of histone H5. Isolation and characterisation of the globular segment". European Journal of Biochemistry / FEBS. 88 (2): 363–71. doi:10.1111/j.1432-1033.1978.tb12457.x. PMID 689022.
- ^ Rizzo PJ (Aug 2003). "Those amazing dinoflagellate chromosomes". Cell Research. 13 (4): 215–7. doi:10.1038/sj.cr.7290166. PMID 12974611.
- ^ Talbert PB, Henikoff S. "Chromatin: Packaging without Nucleosomes". Current Biology. 22 (24): R1040–R1043. doi:10.1016/j.cub.2012.10.052. PMID 23257187.
- ^ Clarke HJ (1992). "Nuclear and chromatin composition of mammalian gametes and early embryos". Biochemistry and Cell Biology = Biochimie Et Biologie Cellulaire. 70 (10–11): 856–66. doi:10.1139/o92-134. PMID 1297351.
- ^ Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L (Dec 2005). "Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning". PLoS Biology. 3 (12): e384. doi:10.1371/journal.pbio.0030384. PMC 1275524. PMID 16248679.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
- ^ Billon P, Côté J (October 2011). "Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance". Biochim Biophys Acta. 1819 (3–4): 290–302. doi:10.1016/j.bbagrm.2011.10.004. PMID 22027408.
- ^ a b Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000). "A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage". Current Biology. 10 (15): 886–95. doi:10.1016/S0960-9822(00)00610-2. PMID 10959836.
- ^ Ahmad K, Henikoff S (Jun 2002). "The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly". Molecular Cell. 9 (6): 1191–200. doi:10.1016/S1097-2765(02)00542-7. PMID 12086617.
- ^ Strahl BD, Allis CD (Jan 2000). "The language of covalent histone modifications". Nature. 403 (6765): 41–5. doi:10.1038/47412. PMID 10638745.
- ^ Jenuwein T, Allis CD (Aug 2001). "Translating the histone code". Science. 293 (5532): 1074–80. doi:10.1126/science.1063127. PMID 11498575.
- ^ Song N, Liu J, An S, Nishino T, Hishikawa Y, Koji T (Aug 2011). "Immunohistochemical Analysis of Histone H3 Modifications in Germ Cells during Mouse Spermatogenesis". Acta Histochemica et Cytochemica. 44 (4): 183–90. doi:10.1267/ahc.11027. PMC 3168764. PMID 21927517.
- ^ Benevolenskaya EV (Aug 2007). "Histone H3K4 demethylases are essential in development and differentiation". Biochemistry and Cell Biology = Biochimie Et Biologie Cellulaire. 85 (4): 435–43. doi:10.1139/o07-057. PMID 17713579.
- ^ a b c d e f g h Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (May 2007). "High-resolution profiling of histone methylations in the human genome". Cell. 129 (4): 823–37. doi:10.1016/j.cell.2007.05.009. PMID 17512414.
- ^ a b c Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR (Apr 2008). "DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells". Molecular and Cellular Biology. 28 (8): 2825–39. doi:10.1128/MCB.02076-07. PMC 2293113. PMID 18285465.
- ^ a b c Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ (2009). "Determination of enriched histone modifications in non-genic portions of the human genome". BMC Genomics. 10: 143. doi:10.1186/1471-2164-10-143. PMC 2667539. PMID 19335899.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
- ^ a b c Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I (Jun 2007). "The landscape of histone modifications across 1% of the human genome in five human cell lines". Genome Research. 17 (6): 691–707. doi:10.1101/gr.5704207. PMC 1891331. PMID 17567990.
- ^ Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (Dec 2010). "Histone H3K27ac separates active from poised enhancers and predicts developmental state". Proceedings of the National Academy of Sciences of the United States of America. 107 (50): 21931–6. doi:10.1073/pnas.1016071107. PMC 3003124. PMID 21106759.
- ^ Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, Nielsen ML, Gurdon JB, Kouzarides T (Mar 2014). "Citrullination regulates pluripotency and histone H1 binding to chromatin". Nature. 507 (7490): 104–8. doi:10.1038/nature12942. PMID 24463520.
- ^ Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T (Sep 2004). "Histone deimination antagonizes arginine methylation". Cell. 118 (5): 545–53. doi:10.1016/j.cell.2004.08.020. PMID 15339660.
- ^ Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (Mar 2003). "The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation". Molecular Cell. 11 (3): 721–9. doi:10.1016/S1097-2765(03)00091-1. PMID 12667454.
- ^ Ng HH, Robert F, Young RA, Struhl K (Mar 2003). "Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity". Molecular Cell. 11 (3): 709–19. doi:10.1016/S1097-2765(03)00092-3. PMID 12667453.
- ^ Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, Schreiber SL, Lander ES (Jan 2005). "Genomic maps and comparative analysis of histone modifications in human and mouse". Cell. 120 (2): 169–81. doi:10.1016/j.cell.2005.01.001. PMID 15680324.
- ^ Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A (Mar 2002). "COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression". The Journal of Biological Chemistry. 277 (13): 10753–5. doi:10.1074/jbc.C200023200. PMID 11805083.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF (Dec 2001). "The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4". The EMBO Journal. 20 (24): 7137–48. doi:10.1093/emboj/20.24.7137. PMC 125774. PMID 11742990.
- ^ Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML (Jan 2002). "A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3". Proceedings of the National Academy of Sciences of the United States of America. 99 (1): 90–4. doi:10.1073/pnas.221596698. PMC 117519. PMID 11752412.
- ^ Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (Nov 2005). "The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS". Molecular Cell. 20 (4): 589–99. doi:10.1016/j.molcel.2005.09.010. PMID 16307922.
- ^ Sarcevic B, Mawson A, Baker RT, Sutherland RL (Apr 2002). "Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation". The EMBO Journal. 21 (8): 2009–18. doi:10.1093/emboj/21.8.2009. PMC 125963. PMID 11953320.
- ^ Robzyk K, Recht J, Osley MA (Jan 2000). "Rad6-dependent ubiquitination of histone H2B in yeast". Science. 287 (5452): 501–4. doi:10.1126/science.287.5452.501. PMID 10642555.
- ^ Sun ZW, Allis CD (Jul 2002). "Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast". Nature. 418 (6893): 104–8. doi:10.1038/nature00883. PMID 12077605.
- ^ Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (Aug 2002). "Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6". The Journal of Biological Chemistry. 277 (32): 28368–71. doi:10.1074/jbc.C200348200. PMID 12070136.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD (Mar 2002). "Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression". Molecular and Cellular Biology. 22 (5): 1298–306. doi:10.1128/MCB.22.5.1298-1306.2002. PMC 134702. PMID 11839797.
- ^ Li J, Moazed D, Gygi SP (Dec 2002). "Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation". The Journal of Biological Chemistry. 277 (51): 49383–8. doi:10.1074/jbc.M209294200. PMID 12381723.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (Nov 2005). "Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription". Cell. 123 (4): 581–92. doi:10.1016/j.cell.2005.10.023. PMID 16286007.
- ^ Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (Nov 2005). "Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex". Cell. 123 (4): 593–605. doi:10.1016/j.cell.2005.10.025. PMID 16286008.
- ^ Joshi AA, Struhl K (Dec 2005). "Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation". Molecular Cell. 20 (6): 971–8. doi:10.1016/j.molcel.2005.11.021. PMID 16364921.
- ^ Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (Nov 2002). "Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein". Genes & Development. 16 (22): 2893–905. doi:10.1101/gad.1035902. PMC 187479. PMID 12435631.
- ^ a b Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (Nov 2002). "Role of histone H3 lysine 27 methylation in Polycomb-group silencing". Science. 298 (5595): 1039–43. doi:10.1126/science.1076997. PMID 12351676.
- ^ de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (Nov 2004). "Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation". Developmental Cell. 7 (5): 663–76. doi:10.1016/j.devcel.2004.10.005. PMID 15525528.
- ^ Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (Oct 2004). "Role of histone H2A ubiquitination in Polycomb silencing". Nature. 431 (7010): 873–8. doi:10.1038/nature02985. PMID 15386022.
- ^ Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N (Feb 2012). "RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3". Cell. 148 (4): 664–78. doi:10.1016/j.cell.2011.12.029. PMC 3281992. PMID 22325148.
- ^ Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D (Feb 2012). "PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes". Molecular Cell. 45 (3): 344–56. doi:10.1016/j.molcel.2012.01.002. PMC 3293217. PMID 22325352.
- ^ Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (Jan 2004). "RNAi-mediated targeting of heterochromatin by the RITS complex". Science. 303 (5658): 672–6. doi:10.1126/science.1093686. PMC 3244756. PMID 14704433.
- ^ Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (Aug 2000). "Regulation of chromatin structure by site-specific histone H3 methyltransferases". Nature. 406 (6796): 593–9. doi:10.1038/35020506. PMID 10949293.
- ^ Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (Mar 2001). "Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain". Nature. 410 (6824): 120–4. doi:10.1038/35065138. PMID 11242054.
- ^ Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (Mar 2001). "Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins". Nature. 410 (6824): 116–20. doi:10.1038/35065132. PMID 11242053.
- ^ a b Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (Jun 2004). "A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin". Genes & Development. 18 (11): 1251–62. doi:10.1101/gad.300704. PMC 420351. PMID 15145825.
- ^ Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Shi W, Fundele R, Singh PB (May 2004). "Heterochromatin and tri-methylated lysine 20 of histone H4 in animals". Journal of Cell Science. 117 (Pt 12): 2491–501. doi:10.1242/jcs.01238. PMID 15128874.
- ^ Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (Apr 2006). "A bivalent chromatin structure marks key developmental genes in embryonic stem cells". Cell. 125 (2): 315–26. doi:10.1016/j.cell.2006.02.041. PMID 16630819.
- ^ a b Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (Mar 1998). "DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139". The Journal of Biological Chemistry. 273 (10): 5858–68. doi:10.1074/jbc.273.10.5858. PMID 9488723.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A (May 2002). "Genomic instability in mice lacking histone H2AX". Science. 296 (5569): 922–7. doi:10.1126/science.1069398. PMID 11934988.
- ^ Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (Oct 2004). "Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break". Current Biology. 14 (19): 1703–11. doi:10.1016/j.cub.2004.09.047. PMID 15458641.
- ^ Rogakou EP, Boon C, Redon C, Bonner WM (Sep 1999). "Megabase chromatin domains involved in DNA double-strand breaks in vivo". The Journal of Cell Biology. 146 (5): 905–16. doi:10.1083/jcb.146.5.905. PMC 2169482. PMID 10477747.
- ^ Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (Feb 2003). "MDC1 is a mediator of the mammalian DNA damage checkpoint". Nature. 421 (6926): 961–6. doi:10.1038/nature01446. PMID 12607005.
- ^ Bekker-Jensen S, Mailand N (Dec 2010). "Assembly and function of DNA double-strand break repair foci in mammalian cells". DNA Repair. 9 (12): 1219–28. doi:10.1016/j.dnarep.2010.09.010. PMID 21035408.
- ^ Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C (Jul 2005). "Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae". The Journal of Biological Chemistry. 280 (28): 25949–52. doi:10.1074/jbc.C500181200. PMID 15888442.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Masumoto H, Hawke D, Kobayashi R, Verreault A (Jul 2005). "A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response". Nature. 436 (7048): 294–8. doi:10.1038/nature03714. PMID 16015338.
- ^ Driscoll R, Hudson A, Jackson SP (Feb 2007). "Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56". Science. 315 (5812): 649–52. doi:10.1126/science.1135862. PMC 3334813. PMID 17272722.
- ^ Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z (Feb 2007). "Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication". Science. 315 (5812): 653–5. doi:10.1126/science.1133234. PMID 17272723.
- ^ Das C, Lucia MS, Hansen KC, Tyler JK (May 2009). "CBP/p300-mediated acetylation of histone H3 on lysine 56". Nature. 459 (7243): 113–7. doi:10.1038/nature07861. PMC 2756583. PMID 19270680.
- ^ Han J, Zhou H, Li Z, Xu RM, Zhang Z (Sep 2007). "Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity". The Journal of Biological Chemistry. 282 (39): 28587–96. doi:10.1074/jbc.M702496200. PMID 17690098.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Wurtele H, Kaiser GS, Bacal J, St-Hilaire E, Lee EH, Tsao S, Dorn J, Maddox P, Lisby M, Pasero P, Verreault A (Jan 2012). "Histone H3 lysine 56 acetylation and the response to DNA replication fork damage". Molecular and Cellular Biology. 32 (1): 154–72. doi:10.1128/MCB.05415-11. PMC 3255698. PMID 22025679.
- ^ Wurtele H, Tsao S, Lépine G, Mullick A, Tremblay J, Drogaris P, Lee EH, Thibault P, Verreault A, Raymond M (Jul 2010). "Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy". Nature Medicine. 16 (7): 774–80. doi:10.1038/nm.2175. PMID 20601951.
- ^ Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W, Neumann H (Jan 2014). "A cascade of histone modifications induces chromatin condensation in mitosis". Science. 343 (6166): 77–80. doi:10.1126/science.1244508. PMID 24385627.
- ^ Johansen KM, Johansen J (2006). "Regulation of chromatin structure by histone H3S10 phosphorylation". Chromosome Research. 14 (4): 393–404. doi:10.1007/s10577-006-1063-4. PMID 16821135.
- ^ Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, García-Muse T, Aguilera A (Nov 2013). "R loops are linked to histone H3 S10 phosphorylation and chromatin condensation". Molecular Cell. 52 (4): 583–90. doi:10.1016/j.molcel.2013.10.006. PMID 24211264.
- ^ Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD (May 2003). "Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase". Cell. 113 (4): 507–17. doi:10.1016/s0092-8674(03)00355-6. PMID 12757711.
- ^ Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD (Jan 2005). "Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae". Cell. 120 (1): 25–36. doi:10.1016/j.cell.2004.11.016. PMID 15652479.
