Human microbiome: Difference between revisions
Citation bot (talk | contribs) Removed parameters. | Use this bot. Report bugs. | Suggested by Jay8g | Category:CS1 maint: PMC embargo expired | #UCB_Category |
m →Nasal cavity: adding italics |
||
| (41 intermediate revisions by 21 users not shown) | |||
| Line 4: | Line 4: | ||
[[File:Skin_Microbiome20169-300.jpg|upright=1.25|thumb|Graphic depicting the human [[skin microbiota]], with relative prevalences of various classes of bacteria]] |
[[File:Skin_Microbiome20169-300.jpg|upright=1.25|thumb|Graphic depicting the human [[skin microbiota]], with relative prevalences of various classes of bacteria]] |
||
The '''human microbiome''' is the aggregate of all [[microbiota]] that reside on or within human |
The '''human microbiome''' is the aggregate of all [[microbiota]] that reside on or within [[human tissue]]s and [[biofluid]]s along with the corresponding [[List of human anatomical features|anatomical sites]] in which they reside,<ref name="pmid26229597" /><ref>{{cite journal |last1=Mousa |first1=Walaa K. |last2=Chehadeh |first2=Fadia |last3=Husband |first3=Shannon |title=Recent Advances in Understanding the Structure and Function of the Human Microbiome |journal=Frontiers in Microbiology |date=2022 |volume=13 |doi=10.3389/fmicb.2022.825338 |pmid=35185849 |doi-access=free |pmc=8851206}}</ref> including the [[human gastrointestinal tract|gastrointestinal tract]], [[Human skin|skin]], [[mammary gland]]s, [[seminal fluid]], [[uterus]], [[ovarian follicle]]s, [[lung]], [[saliva]], [[oral mucosa]], [[conjunctiva]], and the [[biliary tract]]. Types of [[List of human microbiota|human microbiota]] include [[Bacterium|bacteria]], [[archaea]], [[Fungus|fungi]], [[protist]]s, and [[viruses]]. Though [[micro-animal]]s can also live on the human body, they are typically excluded from this definition. In the context of [[genomics]], the term ''human microbiome'' is sometimes used to refer to the collective [[genome]]s of resident microorganisms;<ref name="Prescotts">{{Cite book | vauthors = Sherwood L, Willey J, Woolverton C |url={{google books|plainurl=y|id=sBCSRAAACAAJ}} |title=Prescott's Microbiology |publisher=McGraw Hill |year=2013 |isbn=9780073402406 |edition=9th |location=New York |pages=713–721 |oclc=886600661 |name-list-style=vanc}}</ref> however, the term ''[[metagenome|human metagenome]]'' has the same meaning.<ref name="pmid26229597">{{cite journal | vauthors = Marchesi JR, Ravel J | title = The vocabulary of microbiome research: a proposal | journal = Microbiome | volume = 3 | pages = 31 | date = 2015 | pmid = 26229597 | pmc = 4520061 | doi = 10.1186/s40168-015-0094-5 | quote = <br />Microbiome<br />This term refers to the entire habitat, including the microorganisms (bacteria, archaea, lower and higher eurkaryotes, and viruses), their genomes (i.e., genes), and the surrounding environmental conditions. This definition is based on that of "biome," the biotic and abiotic factors of given environments. Others in the field limit the definition of microbiome to the collection of genes and genomes of members of a microbiota. It is argued that this is the definition of [[Metagenomics|metagenome]], which combined with the environment constitutes the microbiome. | doi-access = free }}</ref> |
||
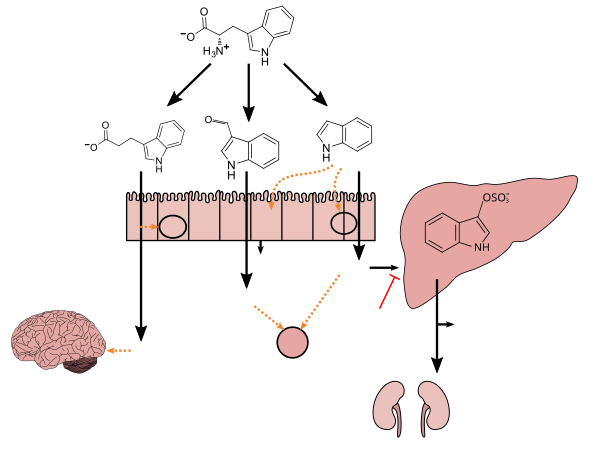
The human body hosts many microorganisms, with approximately the same [[order of magnitude]] of non-human cells as human cells.<ref name="Sender">{{Cite journal |vauthors=Sender R, Fuchs S, Milo R |date=January 2016 |title=Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans |journal=Cell |volume=164 |issue=3 |pages=337–40 |doi=10.1016/j.cell.2016.01.013 |pmid=26824647 |doi-access=free}}</ref> Some microorganisms that humans host are [[commensalism|commensal]], meaning they co-exist without harming humans; others have a [[Mutualism (biology)|mutualistic]] relationship with their human hosts.<ref name="Prescotts" />{{rp|700}}<ref name="Quigley2013rev">{{Cite journal |vauthors=Quigley EM |date=September 2013 |title=Gut bacteria in health and disease |journal=Gastroenterology & Hepatology |volume=9 |issue=9 |pages=560–9 |pmc=3983973 |pmid=24729765}}</ref> Conversely, some non-[[pathogenic]] microorganisms can harm human hosts via the [[metabolites]] they produce, like [[trimethylamine]], which the human body converts to [[trimethylamine N-oxide]] via [[FMO3]]-mediated oxidation.<ref name="TMA">{{Cite journal |vauthors=Falony G, Vieira-Silva S, Raes J |date=2015 |title=Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine |journal=Annual Review of Microbiology |volume=69 |pages=305–21 |doi=10.1146/annurev-micro-091014-104422 |pmid=26274026 |quote=we review literature on trimethylamine (TMA), a microbiota-generated metabolite linked to atherosclerosis development.|doi-access=free }}</ref><ref name="TMA diseases">{{Cite journal |vauthors=Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF |date=November 2014 |title=Archaea and the human gut: new beginning of an old story |journal=World Journal of Gastroenterology |volume=20 |issue=43 |pages=16062–78 |doi=10.3748/wjg.v20.i43.16062 |pmc=4239492 |pmid=25473158 |quote=Trimethylamine is exclusively a microbiota-derived product of nutrients (lecithin, choline, TMAO, L-carnitine) from normal diet, from which seems originate two diseases, trimethylaminuria (or Fish-Odor Syndrome) and cardiovascular disease through the proatherogenic property of its oxidized liver-derived form. |doi-access=free }}</ref> Certain microorganisms perform tasks that are known to be useful to the human host, but the role of most of them is not well understood. Those that are expected to be present, and that under normal circumstances do not cause disease, are sometimes deemed ''normal flora'' or ''normal microbiota''.<ref name="Prescotts" /> |
The human body hosts many microorganisms, with approximately the same [[order of magnitude]] of non-human cells as human cells.<ref name="Sender">{{Cite journal |vauthors=Sender R, Fuchs S, Milo R |date=January 2016 |title=Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans |journal=Cell |volume=164 |issue=3 |pages=337–40 |doi=10.1016/j.cell.2016.01.013 |pmid=26824647 |doi-access=free}}</ref> Some microorganisms that humans host are [[commensalism|commensal]], meaning they co-exist without harming humans; others have a [[Mutualism (biology)|mutualistic]] relationship with their human hosts.<ref name="Prescotts" />{{rp|700}}<ref name="Quigley2013rev">{{Cite journal |vauthors=Quigley EM |date=September 2013 |title=Gut bacteria in health and disease |journal=Gastroenterology & Hepatology |volume=9 |issue=9 |pages=560–9 |pmc=3983973 |pmid=24729765}}</ref> Conversely, some non-[[pathogenic]] microorganisms can harm human hosts via the [[metabolites]] they produce, like [[trimethylamine]], which the human body converts to [[trimethylamine N-oxide]] via [[FMO3]]-mediated oxidation.<ref name="TMA">{{Cite journal |vauthors=Falony G, Vieira-Silva S, Raes J |date=2015 |title=Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine |journal=Annual Review of Microbiology |volume=69 |pages=305–21 |doi=10.1146/annurev-micro-091014-104422 |pmid=26274026 |quote=we review literature on trimethylamine (TMA), a microbiota-generated metabolite linked to atherosclerosis development.|doi-access=free }}</ref><ref name="TMA diseases">{{Cite journal |vauthors=Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF |date=November 2014 |title=Archaea and the human gut: new beginning of an old story |journal=World Journal of Gastroenterology |volume=20 |issue=43 |pages=16062–78 |doi=10.3748/wjg.v20.i43.16062 |pmc=4239492 |pmid=25473158 |quote=Trimethylamine is exclusively a microbiota-derived product of nutrients (lecithin, choline, TMAO, L-carnitine) from normal diet, from which seems originate two diseases, trimethylaminuria (or Fish-Odor Syndrome) and cardiovascular disease through the proatherogenic property of its oxidized liver-derived form. |doi-access=free }}</ref> Certain microorganisms perform tasks that are known to be useful to the human host, but the role of most of them is not well understood. Those that are expected to be present, and that under normal circumstances do not cause disease, are sometimes deemed ''normal flora'' or ''normal microbiota''.<ref name="Prescotts" /> |
||
During early life, the establishment of a diverse and balanced human microbiota plays a critical role in shaping an individual's long-term health.<ref>Smith A, et al. (2019). "The role of the microbiota in the development of allergies and asthma." Current Allergy and Asthma Reports, 19(8), 38.</ref> Studies have shown that the composition of the gut microbiota during infancy is influenced by various factors, including mode of delivery, breastfeeding, and exposure to environmental factors.<ref>Jackson KD, et al. (2016). "Gut microbiota associations with common diseases and prescription medications in a population-based cohort." Nature Communications, 7, 11622.</ref> There are several beneficial species of bacteria and potential probiotics present in [[breast milk]].<ref name="Yi">{{cite journal |vauthors=Yi DY, Kim SY |title=Human Breast Milk Composition and Function in Human Health: From Nutritional Components to Microbiome and MicroRNAs |journal=Nutrients |volume=13 |issue=9 |date=September 2021 |page=3094 |pmid=34578971 |pmc=8471419 |doi=10.3390/nu13093094 |
During early life, the establishment of a diverse and balanced human microbiota plays a critical role in shaping an individual's long-term health.<ref>Smith A, et al. (2019). "The role of the microbiota in the development of allergies and asthma." Current Allergy and Asthma Reports, 19(8), 38.</ref> Studies have shown that the composition of the gut microbiota during infancy is influenced by various factors, including mode of delivery, breastfeeding, and exposure to environmental factors.<ref>Jackson KD, et al. (2016). "Gut microbiota associations with common diseases and prescription medications in a population-based cohort." Nature Communications, 7, 11622.</ref> There are several beneficial species of bacteria and potential probiotics present in [[breast milk]].<ref name="Yi">{{cite journal |vauthors=Yi DY, Kim SY |title=Human Breast Milk Composition and Function in Human Health: From Nutritional Components to Microbiome and MicroRNAs |journal=Nutrients |volume=13 |issue=9 |date=September 2021 |page=3094 |pmid=34578971 |pmc=8471419 |doi=10.3390/nu13093094 |doi-access=free }}</ref> Research has highlighted the beneficial effects of a healthy microbiota in early life, such as the promotion of immune system development, regulation of metabolism, and protection against pathogenic microorganisms.<ref>Yatsunenko T, et al. (2012). "Human gut microbiome viewed across age and geography." Nature, 486(7402), 222-227.</ref> Understanding the complex interplay between the human microbiota and early life health is crucial for developing interventions and strategies to support optimal microbiota development and improve overall health outcomes in individuals.<ref>Sjögren YM, et al. (2009). "Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses." Clinical and Experimental Allergy, 39(12), 1842-1851.</ref> |
||
The [[Human Microbiome Project]] (HMP) took on the project of sequencing the genome of the human microbiota, focusing particularly on the microbiota that normally inhabit the skin, mouth, nose, digestive tract, and vagina.<ref name="Prescotts" /> It reached a milestone in 2012 when it published its initial results.<ref name="hmpdata">{{Cite web |url=http://www.nih.gov/news/health/jun2012/nhgri-13.htm |title=NIH Human Microbiome Project defines normal bacterial makeup of the body |date=13 June 2012 |publisher=NIH News}}</ref> |
The [[Human Microbiome Project]] (HMP) took on the project of sequencing the genome of the human microbiota, focusing particularly on the microbiota that normally inhabit the skin, mouth, nose, digestive tract, and vagina.<ref name="Prescotts" /> It reached a milestone in 2012 when it published its initial results.<ref name="hmpdata">{{Cite web |url=http://www.nih.gov/news/health/jun2012/nhgri-13.htm |title=NIH Human Microbiome Project defines normal bacterial makeup of the body |date=13 June 2012 |publisher=NIH News}}</ref> |
||
| Line 18: | Line 18: | ||
==Relative numbers== |
==Relative numbers== |
||
The number of [[Bacteria cells|bacterial cells]] in the [[human body]] is estimated to be around 38 trillion, while the estimate for [[human cells]] is around 30 trillion.<ref>{{Cite journal |last1=Sender |first1=Ron |last2=Fuchs |first2=Shai |last3=Milo |first3=Ron |date=2016-08-19 |title=Revised Estimates for the Number of Human and Bacteria Cells in the Body |journal=PLOS Biology |language=en |volume=14 |issue=8 |pages=e1002533 |doi=10.1371/journal.pbio.1002533 |issn=1545-7885 |pmc=4991899 |pmid=27541692 |doi-access=free }}</ref><ref>{{Cite journal |last1=Lacy |first1=Brian E. |last2=Spiegel |first2=Brennan |date=July 2019 |title=Introduction to the Gut Microbiome Special Issue |url=https://journals.lww.com/ajg/fulltext/2019/07000/introduction_to_the_gut_microbiome_special_issue.1.aspx |journal= American Journal of Gastroenterology|language=en-US |volume=114 |issue=7 |pages=1013 |doi=10.14309/ajg.0000000000000303 |pmid=31205134 |issn=0002-9270}}</ref><ref>{{Cite journal |last1=Sung |first1=Jaeyun |last2=Rajendraprasad |first2=Sanu S. |last3=Philbrick |first3=Kemuel L. |last4=Bauer |first4=Brent A. |last5=Gajic |first5=Ognjen |last6=Shah |first6=Aditya |last7=Laudanski |first7=Krzysztof |last8=Bakken |first8=Johan S. |last9=Skalski |first9=Joseph |last10=Karnatovskaia |first10=Lioudmila V. |date=February 2024 |title=The human gut microbiome in critical illness: disruptions, consequences, and therapeutic frontiers |journal=Journal of Critical Care |volume=79 |pages=154436 |doi=10.1016/j.jcrc.2023.154436 |issn=1557-8615 |pmid=37769422|doi-access=free |pmc=11034825 }}</ref><ref name="NN2016">{{Cite journal |last=Abbott |first=Alison |date=2016-01-08 |title=Scientists bust myth that our bodies have more bacteria than human cells |url=https://www.nature.com/articles/nature.2016.19136 |journal=Nature |language=en |doi=10.1038/nature.2016.19136 |issn=1476-4687 |s2cid=190879263 |doi-access=free}}</ref> The number of [[bacterial genes]] is estimated to be 2 million, 100 times the number of approximately 20,000 [[human genes]].<ref>{{Cite journal |last1=Gilbert |first1=Jack A. |last2=Blaser |first2=Martin J. |last3=Caporaso |first3=J. Gregory |last4=Jansson |first4=Janet K. |last5=Lynch |first5=Susan V. |last6=Knight |first6=Rob |date=2018-04-10 |title=Current understanding of the human microbiome |journal=Nature Medicine |volume=24 |issue=4 |pages=392–400 |doi=10.1038/nm.4517 |issn=1546-170X |pmc=7043356 |pmid=29634682}}</ref><ref>{{Cite journal |last1=Wischmeyer |first1=Paul E. |last2=McDonald |first2=Daniel |last3=Knight |first3=Rob |date=August 2016 |title=Role of the microbiome, probiotics, and 'dysbiosis therapy' in critical illness |journal=Current Opinion in Critical Care |volume=22 |issue=4 |pages=347–353 |doi=10.1097/MCC.0000000000000321 |issn=1531-7072 |pmc=5065053 |pmid=27327243}}</ref><ref>{{Cite journal |last1=Qin |first1=Junjie |last2=Li |first2=Ruiqiang |last3=Raes |first3=Jeroen |last4=Arumugam |first4=Manimozhiyan |last5=Burgdorf |first5=Kristoffer Solvsten |last6=Manichanh |first6=Chaysavanh |last7=Nielsen |first7=Trine |last8=Pons |first8=Nicolas |last9=Levenez |first9=Florence |last10=Yamada |first10=Takuji |last11=Mende |first11=Daniel R. |last12=Li |first12=Junhua |last13=Xu |first13=Junming |last14=Li |first14=Shaochuan |last15=Li |first15=Dongfang |date=2010-03-04 |title=A human gut microbial gene catalogue established by metagenomic sequencing |journal=Nature |volume=464 |issue=7285 |pages=59–65 |doi=10.1038/nature08821 |issn=1476-4687 |pmc=3779803 |pmid=20203603|bibcode=2010Natur.464...59. }}</ref> |
|||
{{more medical citations needed|section|date=February 2020}} |
|||
Estimates for the number of bacterial cells within the average 70kg Human male is around 38 trillion, with the estimate for human cells around 30 trillion.<ref>{{Cite journal |last1=Sender |first1=Ron |last2=Fuchs |first2=Shai |last3=Milo |first3=Ron |date=2016-08-19 |title=Revised Estimates for the Number of Human and Bacteria Cells in the Body |journal=PLOS Biology |language=en |volume=14 |issue=8 |pages=e1002533 |doi=10.1371/journal.pbio.1002533 |issn=1545-7885 |pmc=4991899 |pmid=27541692 |doi-access=free }}</ref> The number of bacterial genes (assuming 1000 bacterial species in the gut with 2000 genes per species) is estimated to be 2,000,000 genes, 100 times the number of approximately 20,000 human genes.<ref>{{cite journal | vauthors = Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R | title = Current understanding of the human microbiome | journal = Nature Medicine | volume = 24 | issue = 4 | pages = 392–400 | date = April 2018 | pmid = 29634682 | pmc = 7043356 | doi = 10.1038/nm.4517 }}</ref><ref name="NN2016">{{Cite journal|last=Abbott|first=Alison|date=2016-01-08|title=Scientists bust myth that our bodies have more bacteria than human cells|url=https://www.nature.com/articles/nature.2016.19136|journal=Nature|language=en|doi=10.1038/nature.2016.19136|s2cid=190879263 |issn=1476-4687|doi-access=free}}</ref><ref name=Sender/><ref name="AAM2014">American Academy of Microbiology [http://academy.asm.org/index.php/faq-series/5122-humanmicrobiome FAQ: Human Microbiome] {{Webarchive|url=https://web.archive.org/web/20161231092333/http://academy.asm.org/index.php/faq-series/5122-humanmicrobiome |date=31 December 2016 }} January 2014</ref><ref name="Rosner">{{Cite journal|last=Rosner|first=Judah L.|date=2014-02-01|title=Ten Times More Microbial Cells than Body Cells in Humans?|url=http://www.asmscience.org/content/journal/microbe/10.1128/microbe.9.47.2|journal=Microbe Magazine|language=en|volume=9|issue=2|pages=47|doi=10.1128/microbe.9.47.2|issn=1558-7452}}</ref> |
|||
==Study== |
==Study== |
||
| Line 38: | Line 37: | ||
It is a technique that exploits [[Oligonucleotide synthesis|primers]] to target a specific genetic region and enables to determine the microbial [[phylogenies]]. The genetic region is characterized by a highly variable region which can confer detailed identification; it is delimited by conserved regions, which function as [[binding site]]s for primers used in [[Polymerase chain reaction|PCR]]. The main gene used to characterize [[bacteria]] and [[archaea]] is [[16S ribosomal RNA|16S]] [[Ribosomal RNA|rRNA]] gene, while fungi identification is based on [[Internal transcribed spacer|Internal Transcribed Spacer]] (ITS). The technique is fast and not so expensive and enables to obtain a low-resolution classification of a microbial sample; it is optimal for samples that may be contaminated by host DNA. Primer affinity varies among all DNA sequences, which may result in biases during the amplification reaction; indeed, low-abundance samples are susceptible to overamplification errors, since the other contaminating microorganisms result to be over-represented in case of increasing the PCR cycles. Therefore, the optimization of primer selection can help to decrease such errors, although it requires complete knowledge of the microorganisms present in the sample, and their relative abundances.<ref name=":1">{{cite journal | vauthors = Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC | display-authors = 6 | title = Best practices for analysing microbiomes | journal = Nature Reviews. Microbiology | volume = 16 | issue = 7 | pages = 410–422 | date = July 2018 | pmid = 29795328 | doi = 10.1038/s41579-018-0029-9 | s2cid = 43936002 | url = https://biblio.ugent.be/publication/8678762 }}</ref> |
It is a technique that exploits [[Oligonucleotide synthesis|primers]] to target a specific genetic region and enables to determine the microbial [[phylogenies]]. The genetic region is characterized by a highly variable region which can confer detailed identification; it is delimited by conserved regions, which function as [[binding site]]s for primers used in [[Polymerase chain reaction|PCR]]. The main gene used to characterize [[bacteria]] and [[archaea]] is [[16S ribosomal RNA|16S]] [[Ribosomal RNA|rRNA]] gene, while fungi identification is based on [[Internal transcribed spacer|Internal Transcribed Spacer]] (ITS). The technique is fast and not so expensive and enables to obtain a low-resolution classification of a microbial sample; it is optimal for samples that may be contaminated by host DNA. Primer affinity varies among all DNA sequences, which may result in biases during the amplification reaction; indeed, low-abundance samples are susceptible to overamplification errors, since the other contaminating microorganisms result to be over-represented in case of increasing the PCR cycles. Therefore, the optimization of primer selection can help to decrease such errors, although it requires complete knowledge of the microorganisms present in the sample, and their relative abundances.<ref name=":1">{{cite journal | vauthors = Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC | display-authors = 6 | title = Best practices for analysing microbiomes | journal = Nature Reviews. Microbiology | volume = 16 | issue = 7 | pages = 410–422 | date = July 2018 | pmid = 29795328 | doi = 10.1038/s41579-018-0029-9 | s2cid = 43936002 | url = https://biblio.ugent.be/publication/8678762 }}</ref> |
||
Marker gene analysis can be influenced by the primer choice; in this kind of analysis it |
Marker gene analysis can be influenced by the primer choice; in this kind of analysis, it is desirable to use a well-validated protocol (such as the one used in the Earth Microbiome Project). The first thing to do in a marker gene amplicon analysis is to remove sequencing errors; a lot of sequencing platforms are very reliable, but most of the apparent sequence diversity is still due to errors during the sequencing process. To reduce this phenomenon a first approach is to cluster sequences into [[Operational taxonomic unit]] (OTUs): this process consolidates similar sequences (a 97% similarity threshold is usually adopted) into a single feature that can be used in further analysis steps; this method however would discard [[SNPs]] because they would get clustered into a single OTU. Another approach is [[Oligotyping (taxonomy)|Oligotyping]], which includes position-specific information from 16s rRNA sequencing to detect small nucleotide variations and from discriminating between closely related distinct taxa. These methods give as an output a table of DNA sequences and counts of the different sequences per sample rather than OTU.<ref name=":1" /> |
||
Another important step in the analysis is to assign a taxonomic name to microbial sequences in the data. This can be done using [[machine learning]] approaches that can reach an accuracy at genus-level of about 80%. Other popular analysis packages provide support for taxonomic classification using exact matches to reference databases and should provide greater specificity, but poor sensitivity. Unclassified microorganism should be further checked for organelle sequences.<ref name=":1" /> |
Another important step in the analysis is to assign a taxonomic name to microbial sequences in the data. This can be done using [[machine learning]] approaches that can reach an accuracy at genus-level of about 80%. Other popular analysis packages provide support for taxonomic classification using exact matches to reference databases and should provide greater specificity, but poor sensitivity. Unclassified microorganism should be further checked for organelle sequences.<ref name=":1" /> |
||
| Line 58: | Line 57: | ||
[[File:Commensals vs pathogens mechanism.png|thumb|upright=1.25|Commensals vs pathogens mechanism. Mechanisms underlying the inflammation in COPD. Airway epithelium has complex structure: consists of at least seven diverse cell types interacting with each other by means of tight junctions. Moreover, epithelial calls can deliver the signals into the underlying tissues taking part in the mechanisms of innate and adaptive immune defence. The key transmitters of the signals are dendritic cells. Once pathogenic bacterium (e.g., S. pneumoniae, P. aeruginosa) has activated particular pattern recognition receptors on/in epithelial cells, the proinflammatory signaling pathways are activated. This results mainly in IL-1, IL-6 and IL-8 production. These cytokines induce the chemotaxis to the site of infection in its target cells (e.g., neutrophils, dendritic cells and macrophages). On the other hand, representatives of standard microbiota cause only weak signaling preventing the inflammation. The mechanism of distinguishing between harmless and harmful bacteria on the molecular as well as on physiological levels is not completely understood.]] |
[[File:Commensals vs pathogens mechanism.png|thumb|upright=1.25|Commensals vs pathogens mechanism. Mechanisms underlying the inflammation in COPD. Airway epithelium has complex structure: consists of at least seven diverse cell types interacting with each other by means of tight junctions. Moreover, epithelial calls can deliver the signals into the underlying tissues taking part in the mechanisms of innate and adaptive immune defence. The key transmitters of the signals are dendritic cells. Once pathogenic bacterium (e.g., S. pneumoniae, P. aeruginosa) has activated particular pattern recognition receptors on/in epithelial cells, the proinflammatory signaling pathways are activated. This results mainly in IL-1, IL-6 and IL-8 production. These cytokines induce the chemotaxis to the site of infection in its target cells (e.g., neutrophils, dendritic cells and macrophages). On the other hand, representatives of standard microbiota cause only weak signaling preventing the inflammation. The mechanism of distinguishing between harmless and harmful bacteria on the molecular as well as on physiological levels is not completely understood.]] |
||
Populations of microbes (such as [[Bacterium|bacteria]] and [[yeast]]s) inhabit the skin and mucosal surfaces in various parts of the body. Their role forms part of normal, healthy human physiology, however if microbe numbers grow beyond their typical ranges (often due to a compromised immune system) or if microbes populate (such as through poor hygiene or injury) areas of the body normally not colonized or sterile (such as the blood, or the lower respiratory tract, or the abdominal cavity), disease can result (causing, respectively, bacteremia/sepsis, pneumonia, and peritonitis).<ref>{{Cite journal |last1=Kennedy |first1=Katherine M. |last2=de Goffau |first2=Marcus C. |last3=Perez-Muñoz |first3=Maria Elisa |last4=Arrieta |first4=Marie-Claire |last5=Bäckhed |first5=Fredrik |last6=Bork |first6=Peer |last7=Braun |first7=Thorsten |last8=Bushman |first8=Frederic D. |last9=Dore |first9=Joel |last10=de Vos |first10=Willem M. |last11=Earl |first11=Ashlee M. |last12=Eisen |first12=Jonathan A. |last13=Elovitz |first13=Michal A. |last14=Ganal-Vonarburg |first14=Stephanie C. |last15=Gänzle |first15=Michael G. |date=January 2023 |title=Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies |
Populations of microbes (such as [[Bacterium|bacteria]] and [[yeast]]s) inhabit the skin and mucosal surfaces in various parts of the body. Their role forms part of normal, healthy human physiology, however if microbe numbers grow beyond their typical ranges (often due to a compromised immune system) or if microbes populate (such as through poor hygiene or injury) areas of the body normally not colonized or sterile (such as the blood, or the lower respiratory tract, or the abdominal cavity), disease can result (causing, respectively, bacteremia/sepsis, pneumonia, and peritonitis).<ref>{{Cite journal |last1=Kennedy |first1=Katherine M. |last2=de Goffau |first2=Marcus C. |last3=Perez-Muñoz |first3=Maria Elisa |last4=Arrieta |first4=Marie-Claire |last5=Bäckhed |first5=Fredrik |last6=Bork |first6=Peer |last7=Braun |first7=Thorsten |last8=Bushman |first8=Frederic D. |last9=Dore |first9=Joel |last10=de Vos |first10=Willem M. |last11=Earl |first11=Ashlee M. |last12=Eisen |first12=Jonathan A. |last13=Elovitz |first13=Michal A. |last14=Ganal-Vonarburg |first14=Stephanie C. |last15=Gänzle |first15=Michael G. |date=January 2023 |title=Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies |journal=Nature |language=en |volume=613 |issue=7945 |pages=639–649 |doi=10.1038/s41586-022-05546-8 |pmid=36697862 |pmc=11333990 |bibcode=2023Natur.613..639K |s2cid=256268291 |issn=1476-4687|hdl=2164/21298 |hdl-access=free }}</ref> |
||
The Human Microbiome Project found that individuals host thousands of bacterial types, different body sites having their own distinctive communities. Skin and vaginal sites showed smaller diversity than the mouth and gut, these showing the greatest richness. The bacterial makeup for a given site on a body varies from person to person, not only in type, but also in abundance. Bacteria of the same species found throughout the mouth are of multiple subtypes, preferring to inhabit distinctly different locations in the mouth. Even the enterotypes in the human gut, previously thought to be well understood, are from a broad spectrum of communities with blurred taxon boundaries.<ref>[http://www.plos.org/media/press/2012/PLoS_%20HMP_Collection_Manuscript_Summaries.pdf PLoS Human Microbiome Project Collection Manuscript Summaries] {{Webarchive|url=https://web.archive.org/web/20140304161937/http://www.plos.org/media/press/2012/PLoS_%20HMP_Collection_Manuscript_Summaries.pdf |date=4 March 2014 }} 13 June 2012</ref><ref>{{cite web|url = http://www.ucsf.edu/news/2012/06/12149/consortium-scientists-map-human-bodys-bacterial-ecosystem|title = Consortium of Scientists Map the Human Body's Bacterial Ecosystem|work = ucsf.edu| date=13 June 2012 }}</ref> |
The Human Microbiome Project found that individuals host thousands of bacterial types, different body sites having their own distinctive communities. Skin and vaginal sites showed smaller diversity than the mouth and gut, these showing the greatest richness. The bacterial makeup for a given site on a body varies from person to person, not only in type, but also in abundance. Bacteria of the same species found throughout the mouth are of multiple subtypes, preferring to inhabit distinctly different locations in the mouth. Even the enterotypes in the human gut, previously thought to be well understood, are from a broad spectrum of communities with blurred taxon boundaries.<ref>[http://www.plos.org/media/press/2012/PLoS_%20HMP_Collection_Manuscript_Summaries.pdf PLoS Human Microbiome Project Collection Manuscript Summaries] {{Webarchive|url=https://web.archive.org/web/20140304161937/http://www.plos.org/media/press/2012/PLoS_%20HMP_Collection_Manuscript_Summaries.pdf |date=4 March 2014 }} 13 June 2012</ref><ref>{{cite web|url = http://www.ucsf.edu/news/2012/06/12149/consortium-scientists-map-human-bodys-bacterial-ecosystem|title = Consortium of Scientists Map the Human Body's Bacterial Ecosystem|work = ucsf.edu| date=13 June 2012 }}</ref> |
||
| Line 66: | Line 65: | ||
A number of types of bacteria, such as ''[[Actinomyces viscosus]]'' and ''[[Actinomyces naeslundii|A. naeslundii]]'', live in the mouth, where they are part of a sticky substance called [[Dental plaque|plaque]]. If this is not removed by brushing, it hardens into [[Calculus (dental)|calculus]] (also called tartar). The same bacteria also secrete acids that dissolve [[tooth enamel]], causing [[tooth decay]].{{citation needed|date=February 2023}} |
A number of types of bacteria, such as ''[[Actinomyces viscosus]]'' and ''[[Actinomyces naeslundii|A. naeslundii]]'', live in the mouth, where they are part of a sticky substance called [[Dental plaque|plaque]]. If this is not removed by brushing, it hardens into [[Calculus (dental)|calculus]] (also called tartar). The same bacteria also secrete acids that dissolve [[tooth enamel]], causing [[tooth decay]].{{citation needed|date=February 2023}} |
||
The [[Vaginal flora|vaginal microflora]] consist mostly of various [[lactobacillus]] species. It was long thought that the most common of these species was ''[[Lactobacillus acidophilus]]'', but it has later been shown that ''[[Lactobacillus iners|L. iners]]'' is in fact most common, followed by ''[[Lactobacillus crispatus|L. crispatus]]''. Other lactobacilli found in the vagina are ''[[L. jensenii]], [[Lactobacillus delbruekii|L. delbruekii]]'' and ''[[Lactobacillus gasseri|L. gasseri]]''. Disturbance of the vaginal flora can lead to infections such as [[bacterial vaginosis]] and [[ |
The [[Vaginal flora|vaginal microflora]] consist mostly of various [[lactobacillus]] species. It was long thought that the most common of these species was ''[[Lactobacillus acidophilus]]'', but it has later been shown that ''[[Lactobacillus iners|L. iners]]'' is in fact most common, followed by ''[[Lactobacillus crispatus|L. crispatus]]''. Other lactobacilli found in the vagina are ''[[L. jensenii]], [[Lactobacillus delbruekii|L. delbruekii]]'' and ''[[Lactobacillus gasseri|L. gasseri]]''. Disturbance of the vaginal flora can lead to infections such as [[bacterial vaginosis]] and [[candidiasis]].<ref>{{Cite journal |last1=Gajer |first1=Pawel |last2=Brotman |first2=Rebecca M. |last3=Bai |first3=Guoyun |last4=Sakamoto |first4=Joyce |last5=Schütte |first5=Ursel M. E. |last6=Zhong |first6=Xue |last7=Koenig |first7=Sara S. K. |last8=Fu |first8=Li |last9=Ma |first9=Zhanshan (Sam) |last10=Zhou |first10=Xia |last11=Abdo |first11=Zaid |last12=Forney |first12=Larry J. |last13=Ravel |first13=Jacques |date=2012-05-02 |title=Temporal Dynamics of the Human Vaginal Microbiota |journal=Science Translational Medicine |language=en |volume=4 |issue=132 |pages=132ra52 |doi=10.1126/scitranslmed.3003605 |issn=1946-6234 |pmc=3722878 |pmid=22553250}}</ref> |
||
=== Archaea === |
=== Archaea === |
||
[[Archaea]] are present in the human gut, but, in contrast to the enormous variety of [[bacteria]] in this organ, the numbers of archaeal species are much more limited.<ref>{{cite journal | vauthors = Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA | display-authors = 6 | title = Diversity of the human intestinal microbial flora | journal = Science | volume = 308 | issue = 5728 | pages = 1635–8 | date = June 2005 | pmid = 15831718 | pmc = 1395357 | doi = 10.1126/science.1110591 | bibcode = 2005Sci...308.1635E }}</ref> The dominant group are the [[methanogen]]s, particularly ''[[Methanobrevibacter smithii]]'' and ''[[Methanosphaera|Methanosphaera stadtmanae]]''.<ref>{{cite journal | vauthors = Duncan SH, Louis P, Flint HJ | title = Cultivable bacterial diversity from the human colon | journal = Letters in Applied Microbiology | volume = 44 | issue = 4 | pages = 343–50 | date = April 2007 | pmid = 17397470 | doi = 10.1111/j.1472-765X.2007.02129.x | s2cid = 43706882 | doi-access = |
[[Archaea]] are present in the human gut, but, in contrast to the enormous variety of [[bacteria]] in this organ, the numbers of archaeal species are much more limited.<ref>{{cite journal | vauthors = Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA | display-authors = 6 | title = Diversity of the human intestinal microbial flora | journal = Science | volume = 308 | issue = 5728 | pages = 1635–8 | date = June 2005 | pmid = 15831718 | pmc = 1395357 | doi = 10.1126/science.1110591 | bibcode = 2005Sci...308.1635E }}</ref> The dominant group are the [[methanogen]]s, particularly ''[[Methanobrevibacter smithii]]'' and ''[[Methanosphaera|Methanosphaera stadtmanae]]''.<ref>{{cite journal | vauthors = Duncan SH, Louis P, Flint HJ | title = Cultivable bacterial diversity from the human colon | journal = Letters in Applied Microbiology | volume = 44 | issue = 4 | pages = 343–50 | date = April 2007 | pmid = 17397470 | doi = 10.1111/j.1472-765X.2007.02129.x | s2cid = 43706882 | doi-access =}}</ref> However, colonization by methanogens is variable, and only about 50% of humans have easily detectable populations of these organisms.<ref>{{cite journal | vauthors = Florin TH, Zhu G, Kirk KM, Martin NG | title = Shared and unique environmental factors determine the ecology of methanogens in humans and rats | journal = The American Journal of Gastroenterology | volume = 95 | issue = 10 | pages = 2872–9 | date = October 2000 | doi = 10.1111/j.1572-0241.2000.02319.x | pmid = 11051362 | citeseerx = 10.1.1.606.4187 | s2cid = 1087298 }}</ref> |
||
As of 2007, no clear examples of archaeal [[pathogen]]s were known,<ref>{{cite journal | vauthors = Eckburg PB, Lepp PW, Relman DA | title = Archaea and their potential role in human disease | journal = Infection and Immunity | volume = 71 | issue = 2 | pages = 591–6 | date = February 2003 | pmid = 12540534 | pmc = 145348 | doi = 10.1128/IAI.71.2.591-596.2003 }}</ref><ref>{{cite journal | vauthors = Cavicchioli R, Curmi PM, Saunders N, Thomas T | title = Pathogenic archaea: do they exist? | journal = BioEssays | volume = 25 | issue = 11 | pages = 1119–28 | date = November 2003 | pmid = 14579252 | doi = 10.1002/bies.10354 | doi-access = free }}</ref> although a relationship has been proposed between the presence of some methanogens and human [[periodontal disease]].<ref>{{cite journal | vauthors = Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA | title = Methanogenic Archaea and human periodontal disease | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 101 | issue = 16 | pages = 6176–81 | date = April 2004 | pmid = 15067114 | pmc = 395942 | doi = 10.1073/pnas.0308766101 | bibcode = 2004PNAS..101.6176L | doi-access = free }}</ref> |
As of 2007, no clear examples of archaeal [[pathogen]]s were known,<ref>{{cite journal | vauthors = Eckburg PB, Lepp PW, Relman DA | title = Archaea and their potential role in human disease | journal = Infection and Immunity | volume = 71 | issue = 2 | pages = 591–6 | date = February 2003 | pmid = 12540534 | pmc = 145348 | doi = 10.1128/IAI.71.2.591-596.2003 }}</ref><ref>{{cite journal | vauthors = Cavicchioli R, Curmi PM, Saunders N, Thomas T | title = Pathogenic archaea: do they exist? | journal = BioEssays | volume = 25 | issue = 11 | pages = 1119–28 | date = November 2003 | pmid = 14579252 | doi = 10.1002/bies.10354 | doi-access = free }}</ref> although a relationship has been proposed between the presence of some methanogens and human [[periodontal disease]].<ref>{{cite journal | vauthors = Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA | title = Methanogenic Archaea and human periodontal disease | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 101 | issue = 16 | pages = 6176–81 | date = April 2004 | pmid = 15067114 | pmc = 395942 | doi = 10.1073/pnas.0308766101 | bibcode = 2004PNAS..101.6176L | doi-access = free }}</ref> [[Small intestinal bacterial overgrowth#Methane-dominant SIBO|Methane-dominant small intestinal bacterial overgrowth]] (SIBO) is also predominantly caused by methanogens, and ''Methanobrevibacter smithii'' in particular.<ref name=":02">{{Cite journal |last1=Takakura |first1=Will |last2=Pimentel |first2=Mark |date=2020-07-10 |title=Small Intestinal Bacterial Overgrowth and Irritable Bowel Syndrome – An Update |journal=Frontiers in Psychiatry |volume=11 |page=664 |doi=10.3389/fpsyt.2020.00664 |issn=1664-0640 |pmc=7366247 |pmid=32754068 |doi-access=free}}</ref> |
||
=== Fungi === |
=== Fungi === |
||
{{See also|Mycobiota |
{{See also|Mycobiota}} |
||
Fungi, in particular [[yeast]]s, are present in the human gut.<ref name="mycobiome">{{cite journal | vauthors = Cui L, Morris A, Ghedin E | title = The human mycobiome in health and disease | journal = Genome Medicine | volume = 5 | issue = 7 | pages = 63 | date = July 2013 | pmid = 23899327 | pmc = 3978422 | doi = 10.1186/gm467 | doi-access = free }} [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3978422/figure/F2/ Figure 2: Distribution of fungal genera in different body sites]</ref><ref name=Martins2014Candidiasis>{{cite journal | vauthors = Martins N, Ferreira IC, Barros L, Silva S, Henriques M | title = Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment | journal = Mycopathologia | volume = 177 | issue = 5–6 | pages = 223–40 | date = June 2014 | pmid = 24789109 | doi = 10.1007/s11046-014-9749-1 | url = http://repositorium.sdum.uminho.pt/bitstream/1822/31482/1/document_17753_1.pdf | quote = ''Candida'' species and other microorganisms are involved in this complicated fungal infection, but ''Candida albicans'' continues to be the most prevalent. In the past two decades, it has been observed an abnormal overgrowth in the gastrointestinal, urinary and respiratory tracts, not only in immunocompromised patients, but also related to nosocomial infections and even in healthy individuals. There is a widely variety of causal factors that contribute to yeast infection which means that candidiasis is a good example of a multifactorial syndrome. | hdl = 10198/10147 | s2cid = 795450 | hdl-access = free }}</ref><ref name=Wang2014rev>{{cite journal | vauthors = Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH | title = Review article: fungal microbiota and digestive diseases | journal = Alimentary Pharmacology & Therapeutics | volume = 39 | issue = 8 | pages = 751–66 | date = April 2014 | pmid = 24612332 | doi = 10.1111/apt.12665 | s2cid = 22101484 | quote = In addition, GI fungal infection is reported even among those patients with normal immune status. Digestive system-related fungal infections may be induced by both commensal opportunistic fungi and exogenous pathogenic fungi. ... ''Candida'' sp. is also the most frequently identified species among patients with gastric IFI. ... It was once believed that gastric acid could kill microbes entering the stomach and that the unique ecological environment of the stomach was not suitable for microbial colonisation or infection. However, several studies using culture-independent methods confirmed that large numbers of acid-resistant bacteria belonging to eight phyla and up to 120 species exist in the stomach, such as ''Streptococcus'' sp., ''Neisseria'' sp. and ''Lactobacillus'' sp. etc.<sup>26, 27</sup> Furthermore, ''Candida albicans'' can grow well in highly acidic environments,<sup>28</sup> and some genotypes may increase the severity of gastric mucosal lesions.<sup>29</sup> | doi-access = free }}</ref><ref name="SIFO">{{cite journal | vauthors = Erdogan A, Rao SS | title = Small intestinal fungal overgrowth | journal = Current Gastroenterology Reports | volume = 17 | issue = 4 | pages = 16 | date = April 2015 | pmid = 25786900 | doi = 10.1007/s11894-015-0436-2 | s2cid = 3098136 | quote = Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. Two recent studies showed that 26 % (24/94) and 25.3 % (38/150) of a series of patients with unexplained GI symptoms had SIFO. The most common symptoms observed in these patients were belching, bloating, indigestion, nausea, diarrhea, and gas. ... Fungal-bacterial interaction may act in different ways and may either be synergistic or antagonistic or symbiotic [29]. Some bacteria such as ''Lactobacillus'' species can interact and inhibit both the virulence and growth of ''Candida'' species in the gut by producing hydrogen peroxide [30]. Any damage to the mucosal barrier or disruption of GI microbiota with chemotherapy or antibiotic use, inflammatory processes, activation of immune molecules and disruption of epithelial repair may all cause fungal overgrowth [27]. }}</ref> The best-studied of these are ''[[Candida (genus)|Candida]]'' species due to their ability to become pathogenic in [[Immunodeficiency|immunocompromised]] and even in healthy hosts.<ref name=Martins2014Candidiasis/><ref name=Wang2014rev/><ref name="SIFO" /> Yeasts are also present on the skin,<ref name="mycobiome" /> such as ''[[Malassezia]]'' species, where they consume oils secreted from the [[sebaceous gland]]s.<ref>{{cite journal | vauthors = Marcon MJ, Powell DA | title = Human infections due to Malassezia spp | journal = Clinical Microbiology Reviews | volume = 5 | issue = 2 | pages = 101–19 | date = April 1992 | pmid = 1576583 | pmc = 358230 | doi = 10.1128/CMR.5.2.101 }}</ref><ref>{{cite journal | vauthors = Roth RR, James WD | title = Microbial ecology of the skin | journal = Annual Review of Microbiology | volume = 42 | issue = 1 | pages = 441–64 | year = 1988 | pmid = 3144238 | doi = 10.1146/annurev.mi.42.100188.002301 }}</ref> |
Fungi, in particular [[yeast]]s, are present in the human gut.<ref name="mycobiome">{{cite journal | vauthors = Cui L, Morris A, Ghedin E | title = The human mycobiome in health and disease | journal = Genome Medicine | volume = 5 | issue = 7 | pages = 63 | date = July 2013 | pmid = 23899327 | pmc = 3978422 | doi = 10.1186/gm467 | doi-access = free }} [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3978422/figure/F2/ Figure 2: Distribution of fungal genera in different body sites]</ref><ref name=Martins2014Candidiasis>{{cite journal | vauthors = Martins N, Ferreira IC, Barros L, Silva S, Henriques M | title = Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment | journal = Mycopathologia | volume = 177 | issue = 5–6 | pages = 223–40 | date = June 2014 | pmid = 24789109 | doi = 10.1007/s11046-014-9749-1 | url = http://repositorium.sdum.uminho.pt/bitstream/1822/31482/1/document_17753_1.pdf | quote = ''Candida'' species and other microorganisms are involved in this complicated fungal infection, but ''Candida albicans'' continues to be the most prevalent. In the past two decades, it has been observed an abnormal overgrowth in the gastrointestinal, urinary and respiratory tracts, not only in immunocompromised patients, but also related to nosocomial infections and even in healthy individuals. There is a widely variety of causal factors that contribute to yeast infection which means that candidiasis is a good example of a multifactorial syndrome. | hdl = 10198/10147 | s2cid = 795450 | hdl-access = free }}</ref><ref name=Wang2014rev>{{cite journal | vauthors = Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH | title = Review article: fungal microbiota and digestive diseases | journal = Alimentary Pharmacology & Therapeutics | volume = 39 | issue = 8 | pages = 751–66 | date = April 2014 | pmid = 24612332 | doi = 10.1111/apt.12665 | s2cid = 22101484 | quote = In addition, GI fungal infection is reported even among those patients with normal immune status. Digestive system-related fungal infections may be induced by both commensal opportunistic fungi and exogenous pathogenic fungi. ... ''Candida'' sp. is also the most frequently identified species among patients with gastric IFI. ... It was once believed that gastric acid could kill microbes entering the stomach and that the unique ecological environment of the stomach was not suitable for microbial colonisation or infection. However, several studies using culture-independent methods confirmed that large numbers of acid-resistant bacteria belonging to eight phyla and up to 120 species exist in the stomach, such as ''Streptococcus'' sp., ''Neisseria'' sp. and ''Lactobacillus'' sp. etc.<sup>26, 27</sup> Furthermore, ''Candida albicans'' can grow well in highly acidic environments,<sup>28</sup> and some genotypes may increase the severity of gastric mucosal lesions.<sup>29</sup> | doi-access = free }}</ref><ref name="SIFO">{{cite journal | vauthors = Erdogan A, Rao SS | title = Small intestinal fungal overgrowth | journal = Current Gastroenterology Reports | volume = 17 | issue = 4 | pages = 16 | date = April 2015 | pmid = 25786900 | doi = 10.1007/s11894-015-0436-2 | s2cid = 3098136 | quote = Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. Two recent studies showed that 26 % (24/94) and 25.3 % (38/150) of a series of patients with unexplained GI symptoms had SIFO. The most common symptoms observed in these patients were belching, bloating, indigestion, nausea, diarrhea, and gas. ... Fungal-bacterial interaction may act in different ways and may either be synergistic or antagonistic or symbiotic [29]. Some bacteria such as ''Lactobacillus'' species can interact and inhibit both the virulence and growth of ''Candida'' species in the gut by producing hydrogen peroxide [30]. Any damage to the mucosal barrier or disruption of GI microbiota with chemotherapy or antibiotic use, inflammatory processes, activation of immune molecules and disruption of epithelial repair may all cause fungal overgrowth [27]. }}</ref> The best-studied of these are ''[[Candida (genus)|Candida]]'' species due to their ability to become pathogenic in [[Immunodeficiency|immunocompromised]] and even in healthy hosts.<ref name=Martins2014Candidiasis/><ref name=Wang2014rev/><ref name="SIFO" /> Yeasts are also present on the skin,<ref name="mycobiome" /> such as ''[[Malassezia]]'' species, where they consume oils secreted from the [[sebaceous gland]]s.<ref>{{cite journal | vauthors = Marcon MJ, Powell DA | title = Human infections due to Malassezia spp | journal = Clinical Microbiology Reviews | volume = 5 | issue = 2 | pages = 101–19 | date = April 1992 | pmid = 1576583 | pmc = 358230 | doi = 10.1128/CMR.5.2.101 }}</ref><ref>{{cite journal | vauthors = Roth RR, James WD | title = Microbial ecology of the skin | journal = Annual Review of Microbiology | volume = 42 | issue = 1 | pages = 441–64 | year = 1988 | pmid = 3144238 | doi = 10.1146/annurev.mi.42.100188.002301 }}</ref> |
||
| Line 80: | Line 79: | ||
{{See also|Human virome}}Viruses, especially bacterial viruses ([[bacteriophage]]s), colonize various body sites. These colonized sites include the skin,<ref>{{cite journal | vauthors = Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, Minot S, Bushman FD, Grice EA | display-authors = 6 | title = The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome | journal = mBio | volume = 6 | issue = 5 | pages = e01578-15 | date = October 2015 | pmid = 26489866 | pmc = 4620475 | doi = 10.1128/mBio.01578-15 }}</ref> gut,<ref>{{cite journal | vauthors = Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD | display-authors = 6 | title = The human gut virome: inter-individual variation and dynamic response to diet | journal = Genome Research | volume = 21 | issue = 10 | pages = 1616–25 | date = October 2011 | pmid = 21880779 | pmc = 3202279 | doi = 10.1101/gr.122705.111 }}</ref> lungs,<ref>{{cite journal | vauthors = Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, Haas AR, Abbas A, Frye L, Christie JD, Bushman FD, Collman RG | display-authors = 6 | title = Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients | journal = American Journal of Transplantation | volume = 15 | issue = 1 | pages = 200–9 | date = January 2015 | pmid = 25403800 | pmc = 4276431 | doi = 10.1111/ajt.13031 }}</ref> and oral cavity.<ref>{{cite journal | vauthors = Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT | title = Human oral viruses are personal, persistent and gender-consistent | journal = The ISME Journal | volume = 8 | issue = 9 | pages = 1753–67 | date = September 2014 | pmid = 24646696 | pmc = 4139723 | doi = 10.1038/ismej.2014.31 | bibcode = 2014ISMEJ...8.1753A }}</ref> Virus communities have been associated with some diseases, and do not simply reflect the bacterial communities.<ref>{{cite journal | vauthors = Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT | title = Altered oral viral ecology in association with periodontal disease | journal = mBio | volume = 5 | issue = 3 | pages = e01133-14 | date = May 2014 | pmid = 24846382 | pmc = 4030452 | doi = 10.1128/mBio.01133-14 }}</ref><ref>{{cite journal | vauthors = Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, Norman JM, Keller BC, Luévano JM, Wang D, Boum Y, Martin JN, Hunt PW, Bangsberg DR, Siedner MJ, Kwon DS, Virgin HW | display-authors = 6 | title = Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome | journal = Cell Host & Microbe | volume = 19 | issue = 3 | pages = 311–22 | date = March 2016 | pmid = 26962942 | pmc = 4821831 | doi = 10.1016/j.chom.2016.02.011 }}</ref><ref>{{cite journal | vauthors = Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW | display-authors = 6 | title = Disease-specific alterations in the enteric virome in inflammatory bowel disease | journal = Cell | volume = 160 | issue = 3 | pages = 447–60 | date = January 2015 | pmid = 25619688 | pmc = 4312520 | doi = 10.1016/j.cell.2015.01.002 }}</ref> |
{{See also|Human virome}}Viruses, especially bacterial viruses ([[bacteriophage]]s), colonize various body sites. These colonized sites include the skin,<ref>{{cite journal | vauthors = Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, Minot S, Bushman FD, Grice EA | display-authors = 6 | title = The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome | journal = mBio | volume = 6 | issue = 5 | pages = e01578-15 | date = October 2015 | pmid = 26489866 | pmc = 4620475 | doi = 10.1128/mBio.01578-15 }}</ref> gut,<ref>{{cite journal | vauthors = Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD | display-authors = 6 | title = The human gut virome: inter-individual variation and dynamic response to diet | journal = Genome Research | volume = 21 | issue = 10 | pages = 1616–25 | date = October 2011 | pmid = 21880779 | pmc = 3202279 | doi = 10.1101/gr.122705.111 }}</ref> lungs,<ref>{{cite journal | vauthors = Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, Haas AR, Abbas A, Frye L, Christie JD, Bushman FD, Collman RG | display-authors = 6 | title = Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients | journal = American Journal of Transplantation | volume = 15 | issue = 1 | pages = 200–9 | date = January 2015 | pmid = 25403800 | pmc = 4276431 | doi = 10.1111/ajt.13031 }}</ref> and oral cavity.<ref>{{cite journal | vauthors = Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT | title = Human oral viruses are personal, persistent and gender-consistent | journal = The ISME Journal | volume = 8 | issue = 9 | pages = 1753–67 | date = September 2014 | pmid = 24646696 | pmc = 4139723 | doi = 10.1038/ismej.2014.31 | bibcode = 2014ISMEJ...8.1753A }}</ref> Virus communities have been associated with some diseases, and do not simply reflect the bacterial communities.<ref>{{cite journal | vauthors = Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT | title = Altered oral viral ecology in association with periodontal disease | journal = mBio | volume = 5 | issue = 3 | pages = e01133-14 | date = May 2014 | pmid = 24846382 | pmc = 4030452 | doi = 10.1128/mBio.01133-14 }}</ref><ref>{{cite journal | vauthors = Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, Norman JM, Keller BC, Luévano JM, Wang D, Boum Y, Martin JN, Hunt PW, Bangsberg DR, Siedner MJ, Kwon DS, Virgin HW | display-authors = 6 | title = Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome | journal = Cell Host & Microbe | volume = 19 | issue = 3 | pages = 311–22 | date = March 2016 | pmid = 26962942 | pmc = 4821831 | doi = 10.1016/j.chom.2016.02.011 }}</ref><ref>{{cite journal | vauthors = Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW | display-authors = 6 | title = Disease-specific alterations in the enteric virome in inflammatory bowel disease | journal = Cell | volume = 160 | issue = 3 | pages = 447–60 | date = January 2015 | pmid = 25619688 | pmc = 4312520 | doi = 10.1016/j.cell.2015.01.002 }}</ref> |
||
In January 2024, biologists reported the discovery of "[[Obelisk (life form)|obelisks]]", a new class of [[viroid|viroid-like elements]], and "oblins", their related group of proteins, in the human microbiome.<ref name="SA-20240129">{{cite journal |last=Koumoundouros |first=Tessa |title='Obelisks': Entirely New Class of Life Has Been Found in The Human Digestive System |url=https://www.sciencealert.com/obelisks-entirely-new-class-of-life-has-been-found-in-the-human-digestive-system |date=29 January 2024 |journal=[[ScienceAlert]] |url-status=live | |
In January 2024, biologists reported the discovery of "[[Obelisk (life form)|obelisks]]", a new class of [[viroid|viroid-like elements]], and "oblins", their related group of proteins, in the human microbiome.<ref name="SA-20240129">{{cite journal |last=Koumoundouros |first=Tessa |title='Obelisks': Entirely New Class of Life Has Been Found in The Human Digestive System |url=https://www.sciencealert.com/obelisks-entirely-new-class-of-life-has-been-found-in-the-human-digestive-system |date=29 January 2024 |journal=[[ScienceAlert]] |url-status=live |archive-url=https://archive.today/20240129133614/https://www.sciencealert.com/obelisks-entirely-new-class-of-life-has-been-found-in-the-human-digestive-system |archive-date=29 January 2024 |access-date=29 January 2024 }}</ref><ref name="BRX-20240121">{{cite journal |author=Zheludev, Ivan N. |display-authors=et al. |title=Viroid-like colonists of human microbiomes |url=https://www.biorxiv.org/content/10.1101/2024.01.20.576352v1 |date=21 January 2024 |journal=[[bioRxiv]] |pages=2024.01.20.576352 |doi=10.1101/2024.01.20.576352 |pmid=38293115 |url-status=live |archive-url=https://archive.today/20240129133518/https://www.biorxiv.org/content/10.1101/2024.01.20.576352v1 |archive-date=29 January 2024 |access-date=29 January 2024 |pmc=10827157 }}</ref> |
||
== Anatomical areas == |
== Anatomical areas == |
||
| Line 92: | Line 91: | ||
=== Conjunctiva === |
=== Conjunctiva === |
||
A small number of bacteria and fungi are normally present in the [[conjunctiva]].<ref name="mycobiome" /><ref name=Textbook/> Classes of bacteria include Gram-positive cocci (e.g., ''[[Staphylococcus]]'' and [[Streptococcus]]) and Gram-negative rods and cocci (e.g., ''[[Haemophilus]]'' and ''[[Neisseria]]'') are present.<ref name=Textbook>{{cite web|url = http://www.textbookofbacteriology.net/normalflora_3.html|title = The Normal Bacterial Flora of Humans|work = textbookofbacteriology.net}}</ref> Fungal genera include ''Candida'', ''Aspergillus'', and ''Penicillium''.<ref name="mycobiome" /> The [[lachrymal glands]] continuously secrete, keeping the conjunctiva moist, while intermittent blinking lubricates the conjunctiva and washes away foreign material. Tears contain bactericides such as [[lysozyme]], so that microorganisms have difficulty in surviving the lysozyme and settling on the [[Epithelium|epithelial]] surfaces. |
A small number of bacteria and fungi are normally present in the [[conjunctiva]].<ref name="mycobiome" /><ref name=Textbook/> Classes of bacteria include Gram-positive cocci (e.g., ''[[Staphylococcus]]'' and ''[[Streptococcus]]'') and Gram-negative rods and cocci (e.g., ''[[Haemophilus]]'' and ''[[Neisseria]]'') are present.<ref name=Textbook>{{cite web|url = http://www.textbookofbacteriology.net/normalflora_3.html|title = The Normal Bacterial Flora of Humans|work = textbookofbacteriology.net}}</ref> Fungal genera include ''Candida'', ''Aspergillus'', and ''Penicillium''.<ref name="mycobiome" /> The [[lachrymal glands]] continuously secrete, keeping the conjunctiva moist, while intermittent blinking lubricates the conjunctiva and washes away foreign material. Tears contain bactericides such as [[lysozyme]], so that microorganisms have difficulty in surviving the lysozyme and settling on the [[Epithelium|epithelial]] surfaces. |
||
=== Gastrointestinal tract === |
=== Gastrointestinal tract === |
||
| Line 118: | Line 117: | ||
{{Main|Placental microbiome}} |
{{Main|Placental microbiome}} |
||
Until recently the [[placenta]] was considered to be a sterile organ but commensal, nonpathogenic bacterial species and genera have been identified that reside in the placental tissue.<ref name="FoxEichelberger2015">{{cite journal | vauthors = Fox C, Eichelberger K | title = Maternal microbiome and pregnancy outcomes | journal = Fertility and Sterility | volume = 104 | issue = 6 | pages = 1358–63 | date = December 2015 | pmid = 26493119 | doi = 10.1016/j.fertnstert.2015.09.037 | doi-access = free }}</ref><ref name="WassenaarPanigrahi2014">{{cite journal | vauthors = Wassenaar TM, Panigrahi P | title = Is a foetus developing in a sterile environment? | journal = Letters in Applied Microbiology | volume = 59 | issue = 6 | pages = 572–9 | date = December 2014 | pmid = 25273890 | doi = 10.1111/lam.12334 | s2cid = 206169539 | doi-access = |
Until recently the [[placenta]] was considered to be a sterile organ but commensal, nonpathogenic bacterial species and genera have been identified that reside in the placental tissue.<ref name="FoxEichelberger2015">{{cite journal | vauthors = Fox C, Eichelberger K | title = Maternal microbiome and pregnancy outcomes | journal = Fertility and Sterility | volume = 104 | issue = 6 | pages = 1358–63 | date = December 2015 | pmid = 26493119 | doi = 10.1016/j.fertnstert.2015.09.037 | doi-access = free }}</ref><ref name="WassenaarPanigrahi2014">{{cite journal | vauthors = Wassenaar TM, Panigrahi P | title = Is a foetus developing in a sterile environment? | journal = Letters in Applied Microbiology | volume = 59 | issue = 6 | pages = 572–9 | date = December 2014 | pmid = 25273890 | doi = 10.1111/lam.12334 | s2cid = 206169539 | doi-access =}}</ref><ref>{{cite book | last = Schwiertz | first = Andreas | name-list-style = vanc | title = Microbiota of the human body : implications in health and disease | publisher = Springer | location = Switzerland | year = 2016 | page= 1 |isbn = 978-3-319-31248-4 }}</ref> However, the existence of a microbiome in the placenta is controversial as criticized in several researches. So called "placental microbiome" is likely derived from contamination of regents because low-biomass samples are easily contaminated.<ref>{{cite journal | vauthors = Tamburini S, Shen N, Wu HC, Clemente JC | title = The microbiome in early life: implications for health outcomes | journal = Nature Medicine | volume = 22 | issue = 7 | pages = 713–22 | date = July 2016 | pmid = 27387886 | doi = 10.1038/nm.4142 | s2cid = 2462790 | doi-access = free }}</ref><ref>{{cite journal | vauthors = de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GC | display-authors = 6 | title = Human placenta has no microbiome but can contain potential pathogens | journal = Nature | volume = 572 | issue = 7769 | pages = 329–334 | date = August 2019 | pmid = 31367035 | pmc = 6697540 | doi = 10.1038/s41586-019-1451-5 | bibcode = 2019Natur.572..329D }}</ref><ref>{{cite journal | vauthors = Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS | title = Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations | journal = Trends in Microbiology | volume = 27 | issue = 2 | pages = 105–117 | date = February 2019 | pmid = 30497919 | doi = 10.1016/j.tim.2018.11.003 | hdl = 2440/122027 | s2cid = 54166123 | hdl-access = free }}</ref> |
||
===Uterus=== |
===Uterus=== |
||
| Line 129: | Line 128: | ||
The environment present in the human mouth allows the growth of characteristic microorganisms found there. It provides a source of water and nutrients, as well as a moderate temperature.<ref name="Prescotts" /> Resident microbes of the mouth adhere to the teeth and gums to resist mechanical flushing from the mouth to stomach where acid-sensitive microbes are destroyed by [[hydrochloric acid]].<ref name="Prescotts" /><ref name=Wang2014rev/> |
The environment present in the human mouth allows the growth of characteristic microorganisms found there. It provides a source of water and nutrients, as well as a moderate temperature.<ref name="Prescotts" /> Resident microbes of the mouth adhere to the teeth and gums to resist mechanical flushing from the mouth to stomach where acid-sensitive microbes are destroyed by [[hydrochloric acid]].<ref name="Prescotts" /><ref name=Wang2014rev/> |
||
Anaerobic bacteria in the oral cavity include: ''[[Actinomyces]]'', ''[[Arachnia (bacterium)|Arachnia]]'', ''[[Bacteroides]]'', ''[[Bifidobacterium]]'', ''[[Eubacterium]]'', ''[[Fusobacterium]]'', ''[[Lactobacillus]]'', ''[[ |
Anaerobic bacteria in the oral cavity include: ''[[Actinomyces]]'', ''[[Arachnia (bacterium)|Arachnia]]'', ''[[Bacteroides]]'', ''[[Bifidobacterium]]'', ''[[Eubacterium]]'', ''[[Fusobacterium]]'', ''[[Lactobacillus]]'', ''[[Leptotrichia]]'', ''[[Peptococcus]]'', ''[[Peptostreptococcus]]'', ''[[Propionibacterium]]'', ''[[Selenomonas]]'', ''[[Treponema]]'', and ''[[Veillonella]]''.<ref name="sutter">{{cite journal|vauthors=Sutter VL|title=Anaerobes as normal oral flora|journal=Reviews of Infectious Diseases|volume=6|issue=Suppl 1|pages=S62-566|year=1984|pmid= 6372039|doi= 10.1093/clinids/6.Supplement_1.S62}}</ref>{{update inline|date=June 2016}} Genera of fungi that are frequently found in the mouth include ''[[Candida (fungus)|Candida]]'', ''[[Cladosporium]]'', ''[[Aspergillus]]'', ''[[Fusarium]]'', ''[[Glomus (fungus)|Glomus]]'', ''[[Alternaria]]'', ''[[Penicillium]]'', and ''[[Cryptococcus]]'', among others.<ref name="mycobiome" /> |
||
[[Bacteria]] accumulate on both the hard and soft oral tissues in [[biofilm]] allowing them to adhere and strive in the oral environment while protected from the environmental factors and antimicrobial agents.<ref name="systemic disease 4">{{cite journal | vauthors = Kumar PS | title = Oral microbiota and systemic disease | journal = Anaerobe | volume = 24 | pages = 90–3 | date = December 2013 | pmid = 24128801 | doi = 10.1016/j.anaerobe.2013.09.010 | s2cid = 40735283 }}</ref> Saliva plays a key biofilm homeostatic role allowing recolonization of bacteria for formation and controlling growth by detaching biofilm buildup.<ref name="ArweilerNetuschil2016">{{cite book | vauthors = Arweiler NB, Netuschil L |title=Microbiota of the Human Body |series=Advances in Experimental Medicine and Biology |chapter=The Oral Microbiota | veditors = Schwiertz A |volume=902 |date=May 2016 |publisher=Springer, Cham |isbn=978-3-319-31248-4 |pages=45–60 |doi=10.1007/978-3-319-31248-4_4 |pmid=27161350 }}</ref> It also provides a means of nutrients and temperature regulation. The location of the biofilm determines the type of exposed nutrients it receives.<ref name="AvilaOjcius2009">{{cite journal | vauthors = Avila M, Ojcius DM, Yilmaz O | title = The oral microbiota: living with a permanent guest | journal = DNA and Cell Biology | volume = 28 | issue = 8 | pages = 405–11 | date = August 2009 | pmid = 19485767 | pmc = 2768665 | doi = 10.1089/dna.2009.0874 }}</ref> |
[[Bacteria]] accumulate on both the hard and soft oral tissues in [[biofilm]] allowing them to adhere and strive in the oral environment while protected from the environmental factors and antimicrobial agents.<ref name="systemic disease 4">{{cite journal | vauthors = Kumar PS | title = Oral microbiota and systemic disease | journal = Anaerobe | volume = 24 | pages = 90–3 | date = December 2013 | pmid = 24128801 | doi = 10.1016/j.anaerobe.2013.09.010 | s2cid = 40735283 }}</ref> Saliva plays a key biofilm homeostatic role allowing recolonization of bacteria for formation and controlling growth by detaching biofilm buildup.<ref name="ArweilerNetuschil2016">{{cite book | vauthors = Arweiler NB, Netuschil L |title=Microbiota of the Human Body |series=Advances in Experimental Medicine and Biology |chapter=The Oral Microbiota | veditors = Schwiertz A |volume=902 |date=May 2016 |publisher=Springer, Cham |isbn=978-3-319-31248-4 |pages=45–60 |doi=10.1007/978-3-319-31248-4_4 |pmid=27161350 }}</ref> It also provides a means of nutrients and temperature regulation. The location of the biofilm determines the type of exposed nutrients it receives.<ref name="AvilaOjcius2009">{{cite journal | vauthors = Avila M, Ojcius DM, Yilmaz O | title = The oral microbiota: living with a permanent guest | journal = DNA and Cell Biology | volume = 28 | issue = 8 | pages = 405–11 | date = August 2009 | pmid = 19485767 | pmc = 2768665 | doi = 10.1089/dna.2009.0874 }}</ref> |
||
| Line 141: | Line 140: | ||
=== Nasal cavity === |
=== Nasal cavity === |
||
The healthy nasal microbiome is dominated by Corynebacterium |
The healthy nasal microbiome is dominated by ''Corynebacterium'' and ''Staphylococcus'' species. The mucosal microbiome plays a critical role in modulating viral infection.<ref>{{cite journal | vauthors = Rhoades NS, Pinski AN, Monsibais AN, Jankeel A, Doratt BM, Cinco IR, Ibraim I, Messaoudi I | display-authors = 6 | title = Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including ''Pseudomonas aeruginosa'' in the nose | journal = Cell Reports | volume = 36 | issue = 9 | pages = 109637 | date = August 2021 | pmid = 34433082 | pmc = 8361213 | doi = 10.1016/j.celrep.2021.109637|issn=2211-1247 }}</ref> |
||
=== Lung === |
=== Lung === |
||
{{Main|Lung microbiota}} |
{{Main|Lung microbiota}} |
||
Much like the oral cavity, the upper and lower respiratory system possess mechanical deterrents to remove microbes. Goblet cells produce |
Much like the oral cavity, the upper and lower respiratory system possess mechanical deterrents to remove microbes. Goblet cells produce mucus which traps microbes and moves them out of the respiratory system via continuously moving [[Cilium|ciliated]] epithelial cells.<ref name="Prescotts" /> In addition, a bactericidal effect is generated by nasal mucus which contains the enzyme lysozyme.<ref name="Prescotts" /> The upper and lower respiratory tract appears to have its own set of microbiota.<ref name="Man2017">{{Cite journal |author-link3=Debby Bogaert |author1=Wing Ho Man |author2=Wouter A A de Steenhuijsen Piters |author3=Debby Bogaert |doi=10.1038/NRMICRO.2017.14 |issn=1740-1534 | volume = 15 |issue=5 |journal=Nature Reviews Microbiology |language=en |pages=259–270 |pmc=7097736 |pmid=28316330 |publication-date=20 March 2017 |title=The microbiota of the respiratory tract: gatekeeper to respiratory health |year=2017 | hdl = 20.500.11820/f1137874-9c51-401b-bca4-e2a5da3e219b}}[[WDQ (identifier)|Wikidata]]: [[:d:Q34553608|Q34553608]]</ref> Pulmonary bacterial microbiota belong to 9 major bacterial genera: ''Prevotella'', ''Sphingomonas'', ''Pseudomonas'', ''Acinetobacter'', ''Fusobacterium'', ''Megasphaera'', ''Veillonella'', ''Staphylococcus'', and ''Streptococcus''. Some of the bacteria considered "normal biota" in the respiratory tract can cause serious disease especially in immunocompromised individuals; these include ''Streptococcus pyogenes'', ''Haemophilus influenzae'', ''Streptococcus pneumoniae'', ''Neisseria meningitidis'', and ''Staphylococcus aureus''.{{citation needed|date=October 2016}} Fungal genera that compose the pulmonary mycobiome include ''Candida'', ''Malassezia'', ''Neosartorya'', ''Saccharomyces'', and ''Aspergillus'', among others.<ref name="mycobiome" /> |
||
Unusual distributions of bacterial and fungal genera in the respiratory tract is observed in people with [[cystic fibrosis]].<ref name="mycobiome" /><ref name="beringer2000">{{cite journal | vauthors = Beringer PM, Appleman MD | title = Unusual respiratory bacterial flora in cystic fibrosis: microbiologic and clinical features | journal = Current Opinion in Pulmonary Medicine | volume = 6 | issue = 6 | pages = 545–50 | date = November 2000 | pmid = 11100967 | doi = 10.1097/00063198-200011000-00015 | s2cid = 845977 | url = http://www.usc.edu/hsc/pharmacy/pd_labs/COPM11-00.pdf | url-status = dead | archive-url = https://web.archive.org/web/20131016102359/http://www.usc.edu/hsc/pharmacy/pd_labs/COPM11-00.pdf | archive-date = 16 October 2013 }}</ref> Their bacterial flora often contains antibiotic-resistant and slow-growing bacteria, and the frequency of these pathogens changes in relation to age.<ref name="beringer2000" /> |
Unusual distributions of bacterial and fungal genera in the respiratory tract is observed in people with [[cystic fibrosis]].<ref name="mycobiome" /><ref name="beringer2000">{{cite journal | vauthors = Beringer PM, Appleman MD | title = Unusual respiratory bacterial flora in cystic fibrosis: microbiologic and clinical features | journal = Current Opinion in Pulmonary Medicine | volume = 6 | issue = 6 | pages = 545–50 | date = November 2000 | pmid = 11100967 | doi = 10.1097/00063198-200011000-00015 | s2cid = 845977 | url = http://www.usc.edu/hsc/pharmacy/pd_labs/COPM11-00.pdf | url-status = dead | archive-url = https://web.archive.org/web/20131016102359/http://www.usc.edu/hsc/pharmacy/pd_labs/COPM11-00.pdf | archive-date = 16 October 2013 }}</ref> Their bacterial flora often contains antibiotic-resistant and slow-growing bacteria, and the frequency of these pathogens changes in relation to age.<ref name="beringer2000" /> |
||
=== Biliary tract === |
=== Biliary tract === |
||
Traditionally the biliary tract has been considered to be normally sterile, and the presence of microorganisms in bile is a marker of pathological process. This assumption was confirmed by failure in allocation of bacterial strains from the normal bile duct. Papers began emerging in 2013 showing that the normal biliary microbiota is a separate functional layer which protects a biliary tract from colonization by exogenous microorganisms.<ref name=Verdier2015>{{cite journal | vauthors = Verdier J, Luedde T, Sellge G | title = Biliary Mucosal Barrier and Microbiome | journal = Viszeralmedizin | volume = 31 | issue = 3 | pages = 156–61 | date = June 2015 | pmid = 26468308 | pmc = 4569210 | doi = 10.1159/000431071 }}</ref> |
Traditionally the biliary tract has been considered to be normally sterile, and the presence of microorganisms in bile is a marker of pathological process. This assumption was confirmed by failure in allocation of bacterial strains from the normal bile duct. Papers began emerging in 2013 showing that the normal biliary microbiota is a separate functional layer which protects a biliary tract from colonization by exogenous microorganisms.<ref name=Verdier2015>{{cite journal | vauthors = Verdier J, Luedde T, Sellge G | title = Biliary Mucosal Barrier and Microbiome | journal = Viszeralmedizin | volume = 31 | issue = 3 | pages = 156–61 | date = June 2015 | pmid = 26468308 | pmc = 4569210 | doi = 10.1159/000431071 | doi-broken-date = 21 November 2024 }}</ref> |
||
== Disease and death == |
== Disease and death == |
||
Human bodies rely on the innumerable bacterial genes as the source of essential nutrients.<ref>{{cite journal | vauthors = Yu B, Yu B, Yu L | title = Commentary: Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome | journal = Indian Journal of Microbiology | volume = 60 | issue = 2 | pages = 259–261 | date = June 2020 | pmid = 32255860 | pmc = 7105528 | doi = 10.1007/s12088-020-00863-w }}</ref> Both metagenomic and epidemiological studies indicate vital roles for the human microbiome in preventing a wide range of diseases, from type 2 diabetes and obesity to inflammatory bowel disease, Parkinson's disease, and even mental health conditions like depression.<ref>Copeland CS. [http://claudiacopeland.com/uploads/3/5/5/6/35560346/_hjno_the_world_within_us.pdf The World Within Us: Health and the Human Microbiome.] ''Healthcare Journal of New Orleans, Sept-Oct 2017.''</ref> A symbiotic relationship between the [[gut microbiota]] and different bacteria may influence an individual's immune response.<ref>{{cite journal | vauthors = Honda K, Littman DR | title = The microbiota in adaptive immune homeostasis and disease | journal = Nature | volume = 535 | issue = 7610 | pages = 75–84 | date = July 2016 | pmid = 27383982 | doi = 10.1038/nature18848 | s2cid = 4461492 | bibcode = 2016Natur.535...75H }}</ref> Metabolites generated by gut microbes appear to be causative factors in type 2 diabetes.<ref>{{Cite web |title=Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes |url=https://www.metabolon.com/resources/publications/diabetes-serum-metabolites-gut-microbiome/ |access-date=2022-11-03 |website=Metabolon |language=en-US}}</ref> Although in its infancy, microbiome-based treatment is also showing promise, most notably for treating drug-resistant [https://www.vitalacy.com/post/clostridium-difficile-and-hand-hygiene ''C. difficile'']{{ |
Human bodies rely on the innumerable bacterial genes as the source of essential nutrients.<ref>{{cite journal | vauthors = Yu B, Yu B, Yu L | title = Commentary: Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome | journal = Indian Journal of Microbiology | volume = 60 | issue = 2 | pages = 259–261 | date = June 2020 | pmid = 32255860 | pmc = 7105528 | doi = 10.1007/s12088-020-00863-w }}</ref> Both metagenomic and epidemiological studies indicate vital roles for the human microbiome in preventing a wide range of diseases, from type 2 diabetes and obesity to inflammatory bowel disease, Parkinson's disease, and even mental health conditions like depression.<ref>Copeland CS. [http://claudiacopeland.com/uploads/3/5/5/6/35560346/_hjno_the_world_within_us.pdf The World Within Us: Health and the Human Microbiome.] ''Healthcare Journal of New Orleans, Sept-Oct 2017.''</ref> A symbiotic relationship between the [[gut microbiota]] and different bacteria may influence an individual's immune response.<ref>{{cite journal | vauthors = Honda K, Littman DR | title = The microbiota in adaptive immune homeostasis and disease | journal = Nature | volume = 535 | issue = 7610 | pages = 75–84 | date = July 2016 | pmid = 27383982 | doi = 10.1038/nature18848 | s2cid = 4461492 | bibcode = 2016Natur.535...75H }}</ref> Metabolites generated by gut microbes appear to be causative factors in type 2 diabetes.<ref>{{Cite web |title=Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes |url=https://www.metabolon.com/resources/publications/diabetes-serum-metabolites-gut-microbiome/ |access-date=2022-11-03 |website=Metabolon |language=en-US}}</ref> Although in its infancy, microbiome-based treatment is also showing promise, most notably for treating drug-resistant [https://www.vitalacy.com/post/clostridium-difficile-and-hand-hygiene ''C. difficile''] {{Webarchive|url=https://web.archive.org/web/20191207233756/https://www.vitalacy.com/post/clostridium-difficile-and-hand-hygiene |date=7 December 2019 }} infection<ref>{{cite journal | vauthors = Liubakka A, Vaughn BP | title = Clostridium difficile Infection and Fecal Microbiota Transplant | journal = AACN Advanced Critical Care | volume = 27 | issue = 3 | pages = 324–337 | date = July 2016 | pmid = 27959316 | pmc = 5666691 | doi = 10.4037/aacnacc2016703 }}</ref> and in diabetes treatment.<ref>{{cite journal | vauthors = Burton JH, Johnson M, Johnson J, Hsia DS, Greenway FL, Heiman ML | title = Addition of a Gastrointestinal Microbiome Modulator to Metformin Improves Metformin Tolerance and Fasting Glucose Levels | journal = Journal of Diabetes Science and Technology | volume = 9 | issue = 4 | pages = 808–14 | date = July 2015 | pmid = 25802471 | pmc = 4525649 | doi = 10.1177/1932296815577425 }}</ref> |
||
=== ''Clostridioides difficile'' infection === |
=== ''Clostridioides difficile'' infection === |
||
| Line 171: | Line 170: | ||
The [[HIV]] disease progression influences the composition and function of the gut microbiota, with notable differences between HIV-negative, HIV-positive, and post-[[Anti-retroviral|ART]] HIV-positive populations.{{citation needed|date=July 2017}} HIV decreases the integrity of the gut epithelial barrier function by affecting [[tight junction]]s. This breakdown allows for translocation across the gut epithelium, which is thought to contribute to increases in inflammation seen in people with HIV.<ref name=Zilberman2016HIV>{{cite journal | vauthors = Zilberman-Schapira G, Zmora N, Itav S, Bashiardes S, Elinav H, Elinav E |author6-link=Eran Elinav | title = The gut microbiome in human immunodeficiency virus infection | journal = BMC Medicine | volume = 14 | issue = 1 | pages = 83 | date = June 2016 | pmid = 27256449 | pmc = 4891875 | doi = 10.1186/s12916-016-0625-3 |doi-access=free }}</ref> |
The [[HIV]] disease progression influences the composition and function of the gut microbiota, with notable differences between HIV-negative, HIV-positive, and post-[[Anti-retroviral|ART]] HIV-positive populations.{{citation needed|date=July 2017}} HIV decreases the integrity of the gut epithelial barrier function by affecting [[tight junction]]s. This breakdown allows for translocation across the gut epithelium, which is thought to contribute to increases in inflammation seen in people with HIV.<ref name=Zilberman2016HIV>{{cite journal | vauthors = Zilberman-Schapira G, Zmora N, Itav S, Bashiardes S, Elinav H, Elinav E |author6-link=Eran Elinav | title = The gut microbiome in human immunodeficiency virus infection | journal = BMC Medicine | volume = 14 | issue = 1 | pages = 83 | date = June 2016 | pmid = 27256449 | pmc = 4891875 | doi = 10.1186/s12916-016-0625-3 |doi-access=free }}</ref> |
||
Vaginal microbiota plays a role in the infectivity of HIV, with an increased risk of infection and transmission when the woman has [[bacterial vaginosis]], a condition characterized by an abnormal balance of vaginal bacteria.<ref>{{cite journal | vauthors = Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S | title = Vaginal microbiota and its role in HIV transmission and infection | journal = FEMS Microbiology Reviews | volume = 37 | issue = 5 | pages = 762–92 | date = September 2013 | pmid = 23789590 | doi = 10.1111/1574-6976.12029 | s2cid = 32045698 | doi-access = |
Vaginal microbiota plays a role in the infectivity of HIV, with an increased risk of infection and transmission when the woman has [[bacterial vaginosis]], a condition characterized by an abnormal balance of vaginal bacteria.<ref>{{cite journal | vauthors = Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S | title = Vaginal microbiota and its role in HIV transmission and infection | journal = FEMS Microbiology Reviews | volume = 37 | issue = 5 | pages = 762–92 | date = September 2013 | pmid = 23789590 | doi = 10.1111/1574-6976.12029 | s2cid = 32045698 | doi-access =}}</ref> The enhanced infectivity is seen with the increase in [[Proinflammatory cytokine|pro-inflammatory cytokines]] and [[CCR5|CCR5 +]] CD4+ cells in the vagina. However, a decrease in infectivity is seen with increased levels of vaginal ''Lactobacillus,'' which promotes an anti-inflammatory condition.<ref name=Zilberman2016HIV/> |
||
===Gut microbiome of centenarians=== |
===Gut microbiome of centenarians=== |
||
Humans who are 100 years old or older, called [[centenarian]]s, have a distinct gut microbiome. |
Humans who are 100 years old or older, called [[centenarian]]s, have a distinct gut microbiome. This microbiome is characteristically enriched in [[microorganism]]s that are able to synthesize novel secondary [[bile acid]]s.<ref name="Sato2021">Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, Skelly AN, Okamura Y, Vlamakis H, Li Y, Tanoue T, Takei H, Nittono H, Narushima S, Irie J, Itoh H, Moriya K, Sugiura Y, Suematsu M, Moritoki N, Shibata S, Littman DR, Fischbach MA, Uwamino Y, Inoue T, Honda A, Hattori M, Murai T, Xavier RJ, Hirose N, Honda K. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021 Nov;599(7885):458-464. doi: 10.1038/s41586-021-03832-5. Epub 2021 Jul 29. PMID 34325466</ref> These secondary bile acids include various isoforms of [[lithocholic acid]] that may contribute to healthy aging.<ref name = Sato2021/> |
||
=== Death === |
=== Death === |
||
{{main|Necrobiome}} |
{{main|Necrobiome}} |
||
With death, the microbiome of the living body collapses and a different composition of microorganisms named ''[[necrobiome]]'' establishes itself as an important active constituent of the complex physical decomposition process. Its predictable changes over time are thought to be useful to help determine the time of death.<ref name=sa16>{{cite |
With death, the microbiome of the living body collapses and a different composition of microorganisms named ''[[necrobiome]]'' establishes itself as an important active constituent of the complex physical decomposition process. Its predictable changes over time are thought to be useful to help determine the time of death.<ref name=sa16>{{cite magazine |url=https://www.scientificamerican.com/podcast/episode/necrobiome-reveals-a-corpses-time-of-death/ | vauthors = Intagliata C |title="Necrobiome" Reveals a Corpse's Time of Death |magazine=[[Scientific American]] |date=22 December 2016 |access-date=26 March 2018}}</ref><ref name=atl15>{{cite magazine |url= https://www.theatlantic.com/science/archive/2015/12/meet-the-necrobiome-the-predictable-microbes-that-will-eat-your-dying-corpse/419676/ | vauthors = Young E |title= Meet the Necrobiome: The Waves of Microbes That Will Eat Your Corpse |magazine=[[The Atlantic]]| date=10 December 2015| access-date=26 March 2018}}</ref> |
||
== Environmental health == |
== Environmental health == |
||
| Line 186: | Line 185: | ||
== Changes, modulation and transmission == |
== Changes, modulation and transmission == |
||
[[File:Constituents and mechanisms of microbiome-based interventions.webp|thumb|Microbiome-based interventions to modulate gut ecology and the immune system<ref name="10.1038/s41385-022-00564-1"/>]] |
[[File:Constituents and mechanisms of microbiome-based interventions.webp|thumb|Microbiome-based interventions to modulate gut ecology and the immune system<ref name="10.1038/s41385-022-00564-1"/>]] |
||
Hygiene, [[probiotics]],<ref name="10.1038/s41385-022-00564-1">{{cite journal |last1=Hitch |first1=Thomas C. A. |last2=Hall |first2=Lindsay J. |last3=Walsh |first3=Sarah Kate |last4=Leventhal |first4=Gabriel E. |last5=Slack |first5=Emma |last6=de Wouters |first6=Tomas |last7=Walter |first7=Jens |last8=Clavel |first8=Thomas |title=Microbiome-based interventions to modulate gut ecology and the immune system |journal=Mucosal Immunology |date=June 2022 |volume=15 |issue=6 |pages=1095–1113 |doi=10.1038/s41385-022-00564-1 |pmid=36180583 |pmc=9705255 |language=en |issn=1935-3456|doi-access=free}}</ref> [[prebiotics]], microbiota transplants ([[fecal microbiota transplant|fecal]] or skin<ref>{{cite journal |last1=Callewaert |first1=Chris |last2=Knödlseder |first2=Nastassia |last3=Karoglan |first3=Ante |last4=Güell |first4=Marc |last5=Paetzold |first5=Bernhard |title=Skin microbiome transplantation and manipulation: Current state of the art |journal=Computational and Structural Biotechnology Journal |date=1 January 2021 |volume=19 |pages=624–631 |doi=10.1016/j.csbj.2021.01.001 |pmid=33510866 |pmc=7806958 |language=en |issn=2001-0370}}</ref>), |
[[Hygiene]],<ref>{{Cite journal |last1=Yu |first1=Boxuan |last2=Yu |first2=Bowei |last3=Yu |first3=Ligen |date=June 2020 |title=Commentary: Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome |journal=Indian Journal of Microbiology |volume=60 |issue=2 |pages=259–261 |doi=10.1007/s12088-020-00863-w |issn=0046-8991 |pmc=7105528 |pmid=32255860}}</ref> [[probiotics]],<ref name="10.1038/s41385-022-00564-1">{{cite journal |last1=Hitch |first1=Thomas C. A. |last2=Hall |first2=Lindsay J. |last3=Walsh |first3=Sarah Kate |last4=Leventhal |first4=Gabriel E. |last5=Slack |first5=Emma |last6=de Wouters |first6=Tomas |last7=Walter |first7=Jens |last8=Clavel |first8=Thomas |title=Microbiome-based interventions to modulate gut ecology and the immune system |journal=Mucosal Immunology |date=June 2022 |volume=15 |issue=6 |pages=1095–1113 |doi=10.1038/s41385-022-00564-1 |pmid=36180583 |pmc=9705255 |language=en |issn=1935-3456|doi-access=free}}</ref> [[prebiotics]],<ref>{{Cite journal |last1=Roberfroid |first1=Marcel |last2=Gibson |first2=Glenn R. |last3=Hoyles |first3=Lesley |last4=McCartney |first4=Anne L. |last5=Rastall |first5=Robert |last6=Rowland |first6=Ian |last7=Wolvers |first7=Danielle |last8=Watzl |first8=Bernhard |last9=Szajewska |first9=Hania |last10=Stahl |first10=Bernd |last11=Guarner |first11=Francisco |last12=Respondek |first12=Frederique |last13=Whelan |first13=Kevin |last14=Coxam |first14=Veronique |last15=Davicco |first15=Marie-Jeanne |date=August 2010 |title=Prebiotic effects: metabolic and health benefits |url=https://pubmed.ncbi.nlm.nih.gov/20920376/ |journal=The British Journal of Nutrition |volume=104 |issue=Suppl 2 |pages=S1–63 |doi=10.1017/S0007114510003363 |issn=1475-2662 |pmid=20920376}}</ref> [[synbiotics]],<ref>{{Cite journal |last1=Alli |first1=Sauliha R. |last2=Gorbovskaya |first2=Ilona |last3=Liu |first3=Jonathan C. W. |last4=Kolla |first4=Nathan J. |last5=Brown |first5=Lisa |last6=Müller |first6=Daniel J. |date=2022-04-19 |title=The Gut Microbiome in Depression and Potential Benefit of Prebiotics, Probiotics and Synbiotics: A Systematic Review of Clinical Trials and Observational Studies |journal=International Journal of Molecular Sciences |volume=23 |issue=9 |pages=4494 |doi=10.3390/ijms23094494 |doi-access=free |issn=1422-0067 |pmc=9101152 |pmid=35562885}}</ref> [[light therapy]],<ref>{{Cite journal |last1=Ellis |first1=Jeremy R. |last2=Powell |first2=Eron J. |last3=Tomasovic |first3=Luke M. |last4=Marcheskie |first4=Rachel L. |last5=Girish |first5=Vishruth |last6=Warman |first6=Anmol |last7=Sivaloganathan |first7=Darshan |date=June 2024 |title=Changes in the Skin Microbiome Following Dermatological Procedures: A Scoping Review |journal=Applied Microbiology |language=en |volume=4 |issue=2 |pages=972–985 |doi=10.3390/applmicrobiol4020066 |doi-access=free |issn=2673-8007}}</ref> microbiota transplants ([[fecal microbiota transplant|fecal]]<ref>{{Cite journal |last1=Zhou |first1=Siyu |last2=Cui |first2=Ying |last3=Zhang |first3=Yun |last4=Zhao |first4=Tianyu |last5=Cong |first5=Jing |date=2023-03-08 |title=Fecal microbiota transplantation for induction of remission in Crohn's disease: a systematic review and meta-analysis |url=https://pubmed.ncbi.nlm.nih.gov/36882658/ |journal=International Journal of Colorectal Disease |volume=38 |issue=1 |pages=62 |doi=10.1007/s00384-023-04354-4 |issn=1432-1262 |pmid=36882658}}</ref> or [[Skin transplant|skin]]<ref>{{cite journal |last1=Callewaert |first1=Chris |last2=Knödlseder |first2=Nastassia |last3=Karoglan |first3=Ante |last4=Güell |first4=Marc |last5=Paetzold |first5=Bernhard |title=Skin microbiome transplantation and manipulation: Current state of the art |journal=Computational and Structural Biotechnology Journal |date=1 January 2021 |volume=19 |pages=624–631 |doi=10.1016/j.csbj.2021.01.001 |pmid=33510866 |pmc=7806958 |language=en |issn=2001-0370}}</ref>), [[antibiotic]]s,<ref>{{Cite journal |last1=McDonnell |first1=Lucy |last2=Gilkes |first2=Alexander |last3=Ashworth |first3=Mark |last4=Rowland |first4=Victoria |last5=Harries |first5=Timothy Hugh |last6=Armstrong |first6=David |last7=White |first7=Patrick |date=2021 |title=Association between antibiotics and gut microbiome dysbiosis in children: systematic review and meta-analysis |journal=Gut Microbes |volume=13 |issue=1 |pages=1–18 |doi=10.1080/19490976.2020.1870402 |issn=1949-0984 |pmc=7928022 |pmid=33651651}}</ref> [[exercise]],<ref>{{cite journal |last1=Monda |first1=Vincenzo |last2=Villano |first2=Ines |last3=Messina |first3=Antonietta |last4=Valenzano |first4=Anna |last5=Esposito |first5=Teresa |last6=Moscatelli |first6=Fiorenzo |last7=Viggiano |first7=Andrea |last8=Cibelli |first8=Giuseppe |last9=Chieffi |first9=Sergio |last10=Monda |first10=Marcellino |last11=Messina |first11=Giovanni |title=Exercise Modifies the Gut Microbiota with Positive Health Effects |journal=Oxidative Medicine and Cellular Longevity |date=5 March 2017 |volume=2017 |pages=e3831972 |doi=10.1155/2017/3831972 |pmid=28357027 |pmc=5357536 |language=en |issn=1942-0900 |doi-access=free }}</ref><ref>{{cite journal |last1=Mailing |first1=Lucy J. |last2=Allen |first2=Jacob M. |last3=Buford |first3=Thomas W. |last4=Fields |first4=Christopher J. |last5=Woods |first5=Jeffrey A. |title=Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health |journal=Exercise and Sport Sciences Reviews |date=April 2019 |volume=47 |issue=2 |pages=75–85 |doi=10.1249/JES.0000000000000183 |pmid=30883471 |s2cid=83461620 |issn=0091-6331|doi-access=free }}</ref> [[Diet (nutrition)|diet]],<ref>{{Cite journal |last1=So |first1=Daniel |last2=Whelan |first2=Kevin |last3=Rossi |first3=Megan |last4=Morrison |first4=Mark |last5=Holtmann |first5=Gerald |last6=Kelly |first6=Jaimon T. |last7=Shanahan |first7=Erin R. |last8=Staudacher |first8=Heidi M. |last9=Campbell |first9=Katrina L. |date=2018-06-01 |title=Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis |url=https://pubmed.ncbi.nlm.nih.gov/29757343/ |journal=The American Journal of Clinical Nutrition |volume=107 |issue=6 |pages=965–983 |doi=10.1093/ajcn/nqy041 |issn=1938-3207 |pmid=29757343|hdl=10536/DRO/DU:30117355 |hdl-access=free }}</ref> [[breastfeeding]],<ref>{{Cite journal |last1=Ho |first1=Nhan T. |last2=Li |first2=Fan |last3=Lee-Sarwar |first3=Kathleen A. |last4=Tun |first4=Hein M. |last5=Brown |first5=Bryan P. |last6=Pannaraj |first6=Pia S. |last7=Bender |first7=Jeffrey M. |last8=Azad |first8=Meghan B. |last9=Thompson |first9=Amanda L. |last10=Weiss |first10=Scott T. |last11=Azcarate-Peril |first11=M. Andrea |last12=Litonjua |first12=Augusto A. |last13=Kozyrskyj |first13=Anita L. |last14=Jaspan |first14=Heather B. |last15=Aldrovandi |first15=Grace M. |date=2018-10-09 |title=Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations |journal=Nature Communications |volume=9 |issue=1 |pages=4169 |doi=10.1038/s41467-018-06473-x |issn=2041-1723 |pmc=6177445 |pmid=30301893|bibcode=2018NatCo...9.4169H }}</ref> [[senescence|aging]]<ref>{{Cite journal |last1=Lozupone |first1=Catherine A. |last2=Stombaugh |first2=Jesse |last3=Gonzalez |first3=Antonio |last4=Ackermann |first4=Gail |last5=Wendel |first5=Doug |last6=Vázquez-Baeza |first6=Yoshiki |last7=Jansson |first7=Janet K. |last8=Gordon |first8=Jeffrey I. |last9=Knight |first9=Rob |date=October 2013 |title=Meta-analyses of studies of the human microbiota |journal=Genome Research |volume=23 |issue=10 |pages=1704–1714 |doi=10.1101/gr.151803.112 |issn=1549-5469 |pmc=3787266 |pmid=23861384}}</ref> can change the human microbiome across various anatomical systems or regions such as skin and gut. |
||
=== Person-to-person transmission === |
|||
| ⚫ | The human microbiome is transmitted between a [[mother]] and her [[children]], as well as between people living in the same [[household]].<ref>{{cite news |title=Comprehensive profiling of human-to-human oral and intestinal microbial transmission |url=https://www.news-medical.net/news/20230120/Comprehensive-profiling-of-human-to-human-oral-and-intestinal-microbial-transmission.aspx |access-date=16 February 2023 |work=News-Medical.net |date=20 January 2023 |language=en |archive-date=16 February 2023 |archive-url=https://web.archive.org/web/20230216215257/https://www.news-medical.net/news/20230120/Comprehensive-profiling-of-human-to-human-oral-and-intestinal-microbial-transmission.aspx |url-status=live }}</ref><ref>{{cite journal |last1=Valles-Colomer |first1=Mireia |last2=Blanco-Míguez |first2=Aitor |last3=Manghi |first3=Paolo |display-authors=et al.|doi-access=free |title=The person-to-person transmission landscape of the gut and oral microbiomes |journal=Nature |date=February 2023 |volume=614 |issue=7946 |pages=125–135 |doi=10.1038/s41586-022-05620-1 |pmid=36653448 |pmc=9892008 |bibcode=2023Natur.614..125V |language=en |issn=1476-4687}}</ref> |
||
== Research == |
|||
=== Migration === |
=== Migration === |
||
[[Primary research]] indicates that immediate changes in the microbiota may occur when a person [[International migration|migrates]] from one country to another, such as when [[Thailand|Thai]] [[immigrant]]s settled in the United States<ref name="vangay">{{cite journal |display-authors=6 |vauthors=Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, Batres R, Paw B, Pergament SL, Saenyakul P, Xiong M, Kim AD, Kim G, Masopust D, Martens EC, Angkurawaranon C, McGready R, Kashyap PC, Culhane-Pera KA, Knights D |date=November 2018 |title=US Immigration Westernizes the Human Gut Microbiome |journal=Cell |volume=175 |issue=4 |pages=962–972.e10 |doi=10.1016/j.cell.2018.10.029 |pmc=6498444 |pmid=30388453}}</ref> or when [[Latin American]]s immigrated into the United States.<ref name=":3">{{cite journal |display-authors=6 |vauthors=Kaplan RC, Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, Talavera GA, Gellman MD, Thyagarajan B, Moon JY, Vázquez-Baeza Y, McDonald D, Williams-Nguyen JS, Wu MC, North KE, Shaffer J, Sollecito CC, Qi Q, Isasi CR, Wang T, Knight R, Burk RD |date=November 2019 |title=Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity |journal=Genome Biology |volume=20 |issue=1 |pages=219 |doi=10.1186/s13059-019-1831-z |pmc=6824043 |pmid=31672155 |doi-access=free}}</ref> Losses of microbiota diversity were greater in [[obese]] individuals and [[Second generation immigrant|children of immigrants]].<ref name="vangay" /><ref name=":3" /> |
|||
=== |
=== Cellulose digestion === |
||
A 2024 study suggests that [[gut microbiota]] capable of [[Digestion|digesting]] [[cellulose]] can be found in the human microbiome, and they are less abundant in people living in [[Industrialized society|industrialized societies]].<ref>{{Cite journal |last1=Moraïs |first1=Sarah |last2=Winkler |first2=Sarah |last3=Zorea |first3=Alvah |last4=Levin |first4=Liron |last5=Nagies |first5=Falk S. P. |last6=Kapust |first6=Nils |last7=Lamed |first7=Eva |last8=Artan-Furman |first8=Avital |last9=Bolam |first9=David N. |last10=Yadav |first10=Madhav P. |last11=Bayer |first11=Edward A. |last12=Martin |first12=William F. |last13=Mizrahi |first13=Itzhak |date=2024-03-15 |title=Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans |journal=Science |language=en |volume=383 |issue=6688 |pages=eadj9223 |doi=10.1126/science.adj9223 |pmid=38484069 |issn=0036-8075|pmc=7615765 |bibcode=2024Sci...383j9223M }}</ref><ref>{{Cite web |last=Stewart2024-03-14T18:00:00+00:00 |first=Linda |title=Cellulose-degrading gut bacteria found in the human gut, although at lower levels in industrialized countries |url=https://www.the-microbiologist.com/news/cellulose-degrading-gut-bacteria-found-in-the-human-gut-although-at-lower-levels-in-industrialized-countries/2731.article |access-date=2024-03-24 |website=The Microbiologist |language=en}}</ref> |
|||
| ⚫ | |||
== See also == |
== See also == |
||
Latest revision as of 09:23, 10 December 2024

The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside,[1][2] including the gastrointestinal tract, skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, and the biliary tract. Types of human microbiota include bacteria, archaea, fungi, protists, and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term human microbiome is sometimes used to refer to the collective genomes of resident microorganisms;[3] however, the term human metagenome has the same meaning.[1]
The human body hosts many microorganisms, with approximately the same order of magnitude of non-human cells as human cells.[4] Some microorganisms that humans host are commensal, meaning they co-exist without harming humans; others have a mutualistic relationship with their human hosts.[3]: 700 [5] Conversely, some non-pathogenic microorganisms can harm human hosts via the metabolites they produce, like trimethylamine, which the human body converts to trimethylamine N-oxide via FMO3-mediated oxidation.[6][7] Certain microorganisms perform tasks that are known to be useful to the human host, but the role of most of them is not well understood. Those that are expected to be present, and that under normal circumstances do not cause disease, are sometimes deemed normal flora or normal microbiota.[3]
During early life, the establishment of a diverse and balanced human microbiota plays a critical role in shaping an individual's long-term health.[8] Studies have shown that the composition of the gut microbiota during infancy is influenced by various factors, including mode of delivery, breastfeeding, and exposure to environmental factors.[9] There are several beneficial species of bacteria and potential probiotics present in breast milk.[10] Research has highlighted the beneficial effects of a healthy microbiota in early life, such as the promotion of immune system development, regulation of metabolism, and protection against pathogenic microorganisms.[11] Understanding the complex interplay between the human microbiota and early life health is crucial for developing interventions and strategies to support optimal microbiota development and improve overall health outcomes in individuals.[12]
The Human Microbiome Project (HMP) took on the project of sequencing the genome of the human microbiota, focusing particularly on the microbiota that normally inhabit the skin, mouth, nose, digestive tract, and vagina.[3] It reached a milestone in 2012 when it published its initial results.[13]
Terminology
[edit]−4500 — – — – −4000 — – — – −3500 — – — – −3000 — – — – −2500 — – — – −2000 — – — – −1500 — – — – −1000 — – — – −500 — – — – 0 — |
| |||||||||||||||||||||||||||||||||||||||||||||
Though widely known as flora or microflora, this is a misnomer in technical terms, since the word root flora pertains to plants, and biota refers to the total collection of organisms in a particular ecosystem. Recently, the more appropriate term microbiota is applied, though its use has not eclipsed the entrenched use and recognition of flora with regard to bacteria and other microorganisms. Both terms are being used in different literature.[5]
Relative numbers
[edit]The number of bacterial cells in the human body is estimated to be around 38 trillion, while the estimate for human cells is around 30 trillion.[14][15][16][17] The number of bacterial genes is estimated to be 2 million, 100 times the number of approximately 20,000 human genes.[18][19][20]
Study
[edit]
The problem of elucidating the human microbiome is essentially identifying the members of a microbial community, which includes bacteria, eukaryotes, and viruses.[21] This is done primarily using deoxyribonucleic acid (DNA)-based studies, though ribonucleic acid (RNA), protein and metabolite based studies are also performed.[21][22] DNA-based microbiome studies typically can be categorized as either targeted amplicon studies or, more recently, shotgun metagenomic studies. The former focuses on specific known marker genes and is primarily informative taxonomically, while the latter is an entire metagenomic approach which can also be used to study the functional potential of the community.[21] One of the challenges that is present in human microbiome studies, but not in other metagenomic studies, is to avoid including the host DNA in the study.[23]
Aside from simply elucidating the composition of the human microbiome, one of the major questions involving the human microbiome is whether there is a "core", that is, whether there is a subset of the community that is shared among most humans.[24][25] If there is a core, then it would be possible to associate certain community compositions with disease states, which is one of the goals of the HMP. It is known that the human microbiome (such as the gut microbiota) is highly variable both within a single subject and among different individuals, a phenomenon which is also observed in mice.[5]
On 13 June 2012, a major milestone of the HMP was announced by the National Institutes of Health (NIH) director Francis Collins.[13] The announcement was accompanied with a series of coordinated articles published in Nature[26][27] and several journals in the Public Library of Science (PLoS) on the same day. By mapping the normal microbial make-up of healthy humans using genome sequencing techniques, the researchers of the HMP have created a reference database and the boundaries of normal microbial variation in humans. From 242 healthy U.S. volunteers, more than 5,000 samples were collected from tissues from 15 (men) to 18 (women) body sites such as mouth, nose, skin, lower intestine (stool), and vagina. All the DNA, human and microbial, were analyzed with DNA sequencing machines. The microbial genome data were extracted by identifying the bacterial specific ribosomal RNA, 16S rRNA. The researchers calculated that more than 10,000 microbial species occupy the human ecosystem, and they have identified 81–99% of the genera.[28]
Analysis after the processing
[edit]The statistical analysis is essential to validate the obtained results (ANOVA can be used to size the differences between the groups); if it is paired with graphical tools, the outcome is easily visualized and understood.[29]
Once a metagenome is assembled, it is possible to infer the functional potential of the microbiome. The computational challenges for this type of analysis are greater than for single genomes, because usually metagenomes assemblers have poorer quality, and many recovered genes are non-complete or fragmented. After the gene identification step, the data can be used to carry out a functional annotation by means of multiple alignment of the target genes against orthologs databases.[30]
Marker gene analysis
[edit]It is a technique that exploits primers to target a specific genetic region and enables to determine the microbial phylogenies. The genetic region is characterized by a highly variable region which can confer detailed identification; it is delimited by conserved regions, which function as binding sites for primers used in PCR. The main gene used to characterize bacteria and archaea is 16S rRNA gene, while fungi identification is based on Internal Transcribed Spacer (ITS). The technique is fast and not so expensive and enables to obtain a low-resolution classification of a microbial sample; it is optimal for samples that may be contaminated by host DNA. Primer affinity varies among all DNA sequences, which may result in biases during the amplification reaction; indeed, low-abundance samples are susceptible to overamplification errors, since the other contaminating microorganisms result to be over-represented in case of increasing the PCR cycles. Therefore, the optimization of primer selection can help to decrease such errors, although it requires complete knowledge of the microorganisms present in the sample, and their relative abundances.[31]
Marker gene analysis can be influenced by the primer choice; in this kind of analysis, it is desirable to use a well-validated protocol (such as the one used in the Earth Microbiome Project). The first thing to do in a marker gene amplicon analysis is to remove sequencing errors; a lot of sequencing platforms are very reliable, but most of the apparent sequence diversity is still due to errors during the sequencing process. To reduce this phenomenon a first approach is to cluster sequences into Operational taxonomic unit (OTUs): this process consolidates similar sequences (a 97% similarity threshold is usually adopted) into a single feature that can be used in further analysis steps; this method however would discard SNPs because they would get clustered into a single OTU. Another approach is Oligotyping, which includes position-specific information from 16s rRNA sequencing to detect small nucleotide variations and from discriminating between closely related distinct taxa. These methods give as an output a table of DNA sequences and counts of the different sequences per sample rather than OTU.[31]
Another important step in the analysis is to assign a taxonomic name to microbial sequences in the data. This can be done using machine learning approaches that can reach an accuracy at genus-level of about 80%. Other popular analysis packages provide support for taxonomic classification using exact matches to reference databases and should provide greater specificity, but poor sensitivity. Unclassified microorganism should be further checked for organelle sequences.[31]
Phylogenetic analysis
[edit]Many methods that exploit phylogenetic inference use the 16SRNA gene for Archea and Bacteria and the 18SRNA gene for Eukaryotes. Phylogenetic comparative methods (PCS) are based on the comparison of multiple traits among microorganisms; the principle is: the closely they are related, the higher number of traits they share. Usually PCS are coupled with phylogenetic generalized least square (PGLS) or other statistical analysis to get more significant results. Ancestral state reconstruction is used in microbiome studies to impute trait values for taxa whose traits are unknown. This is commonly performed with PICRUSt, which relies on available databases. Phylogenetic variables are chosen by researchers according to the type of study: through the selection of some variables with significant biological informations, it is possible to reduce the dimension of the data to analyse.[32]
Phylogenetic aware distance is usually performed with UniFrac or similar tools, such as Soresen's index or Rao's D, to quantify the differences between the different communities. All this methods are negatively affected by horizontal gene transmission (HGT), since it can generate errors and lead to the correlation of distant species. There are different ways to reduce the negative impact of HGT: the use of multiple genes or computational tools to assess the probability of putative HGT events.[32]
Ecological Network analysis
[edit]Microbial communities develop in a very complex dynamic which can be viewed and analyzed as an ecosystem. The ecological interactions between microbes govern its change, equilibrium and stability, and can be represented by a population dynamic model.[33] The ongoing study of ecological features of the microbiome is growing rapidly and allows to understand the fundamental properties of the microbiome. Understanding the underlying rules of microbial community could help with treating diseases related to unstable microbial communities. A very basic question is if different humans, who share different microbial communities, have the same underlying microbial dynamics.[34] Increasing evidence and indications have found that the dynamics is indeed universal.[35] This question is a basic step that will allow scientists to develop treatment strategies, based on the complex dynamics of human microbial communities. There are more important properties on which considerations should be taken into account for developing interventions strategies for controlling the human microbial dynamics.[36] Controlling the microbial communities could result in solving very bad and harmful diseases.
Types
[edit]Bacteria
[edit]
Populations of microbes (such as bacteria and yeasts) inhabit the skin and mucosal surfaces in various parts of the body. Their role forms part of normal, healthy human physiology, however if microbe numbers grow beyond their typical ranges (often due to a compromised immune system) or if microbes populate (such as through poor hygiene or injury) areas of the body normally not colonized or sterile (such as the blood, or the lower respiratory tract, or the abdominal cavity), disease can result (causing, respectively, bacteremia/sepsis, pneumonia, and peritonitis).[37]
The Human Microbiome Project found that individuals host thousands of bacterial types, different body sites having their own distinctive communities. Skin and vaginal sites showed smaller diversity than the mouth and gut, these showing the greatest richness. The bacterial makeup for a given site on a body varies from person to person, not only in type, but also in abundance. Bacteria of the same species found throughout the mouth are of multiple subtypes, preferring to inhabit distinctly different locations in the mouth. Even the enterotypes in the human gut, previously thought to be well understood, are from a broad spectrum of communities with blurred taxon boundaries.[38][39]
It is estimated that 500 to 1,000 species of bacteria live in the human gut but belong to just a few phyla: Bacillota and Bacteroidota dominate but there are also Pseudomonadota, Verrucomicrobiota, Actinobacteriota, Fusobacteriota, and "Cyanobacteria".[40]
A number of types of bacteria, such as Actinomyces viscosus and A. naeslundii, live in the mouth, where they are part of a sticky substance called plaque. If this is not removed by brushing, it hardens into calculus (also called tartar). The same bacteria also secrete acids that dissolve tooth enamel, causing tooth decay.[citation needed]
The vaginal microflora consist mostly of various lactobacillus species. It was long thought that the most common of these species was Lactobacillus acidophilus, but it has later been shown that L. iners is in fact most common, followed by L. crispatus. Other lactobacilli found in the vagina are L. jensenii, L. delbruekii and L. gasseri. Disturbance of the vaginal flora can lead to infections such as bacterial vaginosis and candidiasis.[41]
Archaea
[edit]Archaea are present in the human gut, but, in contrast to the enormous variety of bacteria in this organ, the numbers of archaeal species are much more limited.[42] The dominant group are the methanogens, particularly Methanobrevibacter smithii and Methanosphaera stadtmanae.[43] However, colonization by methanogens is variable, and only about 50% of humans have easily detectable populations of these organisms.[44]
As of 2007, no clear examples of archaeal pathogens were known,[45][46] although a relationship has been proposed between the presence of some methanogens and human periodontal disease.[47] Methane-dominant small intestinal bacterial overgrowth (SIBO) is also predominantly caused by methanogens, and Methanobrevibacter smithii in particular.[48]
Fungi
[edit]Fungi, in particular yeasts, are present in the human gut.[49][50][51][52] The best-studied of these are Candida species due to their ability to become pathogenic in immunocompromised and even in healthy hosts.[50][51][52] Yeasts are also present on the skin,[49] such as Malassezia species, where they consume oils secreted from the sebaceous glands.[53][54]
Viruses
[edit]Viruses, especially bacterial viruses (bacteriophages), colonize various body sites. These colonized sites include the skin,[55] gut,[56] lungs,[57] and oral cavity.[58] Virus communities have been associated with some diseases, and do not simply reflect the bacterial communities.[59][60][61]
In January 2024, biologists reported the discovery of "obelisks", a new class of viroid-like elements, and "oblins", their related group of proteins, in the human microbiome.[62][63]
Anatomical areas
[edit]Skin
[edit]A study of 20 skin sites on each of ten healthy humans found 205 identified genera in 19 bacterial phyla, with most sequences assigned to four phyla: Actinomycetota (51.8%), Bacillota (24.4%), Pseudomonadota (16.5%), and Bacteroidota (6.3%).[64] A large number of fungal genera are present on healthy human skin, with some variability by region of the body; however, during pathological conditions, certain genera tend to dominate in the affected region.[49] For example, Malassezia is dominant in atopic dermatitis and Acremonium is dominant on dandruff-affected scalps.[49]
The skin acts as a barrier to deter the invasion of pathogenic microbes. The human skin contains microbes that reside either in or on the skin and can be residential or transient. Resident microorganism types vary in relation to skin type on the human body. A majority of microbes reside on superficial cells on the skin or prefer to associate with glands. These glands such as oil or sweat glands provide the microbes with water, amino acids, and fatty acids. In addition, resident bacteria that associated with oil glands are often Gram-positive and can be pathogenic.[3]
Conjunctiva
[edit]A small number of bacteria and fungi are normally present in the conjunctiva.[49][65] Classes of bacteria include Gram-positive cocci (e.g., Staphylococcus and Streptococcus) and Gram-negative rods and cocci (e.g., Haemophilus and Neisseria) are present.[65] Fungal genera include Candida, Aspergillus, and Penicillium.[49] The lachrymal glands continuously secrete, keeping the conjunctiva moist, while intermittent blinking lubricates the conjunctiva and washes away foreign material. Tears contain bactericides such as lysozyme, so that microorganisms have difficulty in surviving the lysozyme and settling on the epithelial surfaces.
Gastrointestinal tract
[edit]Tryptophan metabolism by human gut microbiota ()
|
In humans, the composition of the gastrointestinal microbiome is established during birth.[70] Birth by Cesarean section or vaginal delivery also influences the gut's microbial composition. Babies born through the vaginal canal have non-pathogenic, beneficial gut microbiota similar to those found in the mother.[71] However, the gut microbiota of babies delivered by C-section harbors more pathogenic bacteria such as Escherichia coli and Staphylococcus and it takes longer to develop non-pathogenic, beneficial gut microbiota.[72]
The relationship between some gut microbiota and humans is not merely commensal (a non-harmful coexistence), but rather a mutualistic relationship.[3] Some human gut microorganisms benefit the host by fermenting dietary fiber into short-chain fatty acids (SCFAs), such as acetic acid and butyric acid, which are then absorbed by the host.[5][73] Intestinal bacteria also play a role in synthesizing vitamin B and vitamin K as well as metabolizing bile acids, sterols, and xenobiotics.[3][73] The systemic importance of the SCFAs and other compounds they produce are like hormones and the gut flora itself appears to function like an endocrine organ,[73] and dysregulation of the gut flora has been correlated with a host of inflammatory and autoimmune conditions.[5][74]
The composition of human gut microbiota changes over time, when the diet changes, and as overall health changes.[5][74] A systematic review of 15 human randomized controlled trials from July 2016 found that certain commercially available strains of probiotic bacteria from the Bifidobacterium and Lactobacillus genera (B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei), when taken by mouth in daily doses of 109–1010 colony forming units (CFU) for 1–2 months, possess treatment efficacy (i.e., improves behavioral outcomes) in certain central nervous system disorders – including anxiety, depression, autism spectrum disorder, and obsessive–compulsive disorder – and improves certain aspects of memory.[75]
Urethra and bladder
[edit]The genitourinary system appears to have a microbiota,[76][77] which is an unexpected finding in light of the long-standing use of standard clinical microbiological culture methods to detect bacteria in urine when people show signs of a urinary tract infection; it is common for these tests to show no bacteria present.[78] It appears that common culture methods do not detect many kinds of bacteria and other microorganisms that are normally present.[78] As of 2017, sequencing methods were used to identify these microorganisms to determine if there are differences in microbiota between people with urinary tract problems and those who are healthy.[76][77][79] To properly assess the microbiome of the bladder as opposed to the genitourinary system, the urine specimen should be collected directly from the bladder, which is often done with a catheter.[80]
Vagina
[edit]Vaginal microbiota refers to those species and genera that colonize the vagina. These organisms play an important role in protecting against infections and maintaining vaginal health.[81] The most abundant vaginal microorganisms found in premenopausal women are from the genus Lactobacillus, which suppress pathogens by producing hydrogen peroxide and lactic acid.[51][81][82] Bacterial species composition and ratios vary depending on the stage of the menstrual cycle.[83][84][needs update] Ethnicity also influences vaginal flora. The occurrence of hydrogen peroxide-producing lactobacilli is lower in African American women and vaginal pH is higher.[85] Other influential factors such as sexual intercourse and antibiotics have been linked to the loss of lactobacilli.[82] Moreover, studies have found that sexual intercourse with a condom does appear to change lactobacilli levels, and does increase the level of Escherichia coli within the vaginal flora.[82] Changes in the normal, healthy vaginal microbiota is an indication of infections,[86] such as candidiasis or bacterial vaginosis.[82] Candida albicans inhibits the growth of Lactobacillus species, while Lactobacillus species which produce hydrogen peroxide inhibit the growth and virulence of Candida albicans in both the vagina and the gut.[49][51][52]
Fungal genera that have been detected in the vagina include Candida, Pichia, Eurotium, Alternaria, Rhodotorula, and Cladosporium, among others.[49]
Placenta
[edit]Until recently the placenta was considered to be a sterile organ but commensal, nonpathogenic bacterial species and genera have been identified that reside in the placental tissue.[87][88][89] However, the existence of a microbiome in the placenta is controversial as criticized in several researches. So called "placental microbiome" is likely derived from contamination of regents because low-biomass samples are easily contaminated.[90][91][92]
Uterus
[edit]Until recently, the upper reproductive tract of women was considered to be a sterile environment. A variety of microorganisms inhabit the uterus of healthy, asymptomatic women of reproductive age. The microbiome of the uterus differs significantly from that of the vagina and gastrointestinal tract.[93]
Oral cavity
[edit]The environment present in the human mouth allows the growth of characteristic microorganisms found there. It provides a source of water and nutrients, as well as a moderate temperature.[3] Resident microbes of the mouth adhere to the teeth and gums to resist mechanical flushing from the mouth to stomach where acid-sensitive microbes are destroyed by hydrochloric acid.[3][51]
Anaerobic bacteria in the oral cavity include: Actinomyces, Arachnia, Bacteroides, Bifidobacterium, Eubacterium, Fusobacterium, Lactobacillus, Leptotrichia, Peptococcus, Peptostreptococcus, Propionibacterium, Selenomonas, Treponema, and Veillonella.[94][needs update] Genera of fungi that are frequently found in the mouth include Candida, Cladosporium, Aspergillus, Fusarium, Glomus, Alternaria, Penicillium, and Cryptococcus, among others.[49]
Bacteria accumulate on both the hard and soft oral tissues in biofilm allowing them to adhere and strive in the oral environment while protected from the environmental factors and antimicrobial agents.[95] Saliva plays a key biofilm homeostatic role allowing recolonization of bacteria for formation and controlling growth by detaching biofilm buildup.[96] It also provides a means of nutrients and temperature regulation. The location of the biofilm determines the type of exposed nutrients it receives.[97]
Oral bacteria have evolved mechanisms to sense their environment and evade or modify the host. However, a highly efficient innate host defense system constantly monitors the bacterial colonization and prevents bacterial invasion of local tissues. A dynamic equilibrium exists between dental plaque bacteria and the innate host defense system.[98]
This dynamic between host oral cavity and oral microbes plays a key role in health and disease as it provides entry into the body.[99] A healthy equilibrium presents a symbiotic relationship where oral microbes limit growth and adherence of pathogens while the host provides an environment for them to flourish.[99][95] Ecological changes such as change of immune status, shift of resident microbes and nutrient availability shift from a mutual to parasitic relationship resulting in the host being prone to oral and systemic disease.[95] Systemic diseases such as diabetes and cardiovascular diseases has been correlated to poor oral health.[99] Of particular interest is the role of oral microorganisms in the two major dental diseases: dental caries and periodontal disease.[98] Pathogen colonization at the periodontium cause an excessive immune response resulting in a periodontal pocket- a deepened space between the tooth and gingiva.[95] This acts as a protected blood-rich reservoir with nutrients for anaerobic pathogens.[95] Systemic disease at various sites of the body can result from oral microbes entering the blood bypassing periodontal pockets and oral membranes.[99]
Persistent proper oral hygiene is the primary method for preventing oral and systemic disease.[99] It reduces the density of biofilm and overgrowth of potential pathogenic bacteria resulting in disease.[97] However, proper oral hygiene may not be enough as the oral microbiome, genetics, and changes to immune response play a factor in developing chronic infections.[97] Use of antibiotics could treat already spreading infection but ineffective against bacteria within biofilms.[97]
Nasal cavity
[edit]The healthy nasal microbiome is dominated by Corynebacterium and Staphylococcus species. The mucosal microbiome plays a critical role in modulating viral infection.[100]
Lung
[edit]Much like the oral cavity, the upper and lower respiratory system possess mechanical deterrents to remove microbes. Goblet cells produce mucus which traps microbes and moves them out of the respiratory system via continuously moving ciliated epithelial cells.[3] In addition, a bactericidal effect is generated by nasal mucus which contains the enzyme lysozyme.[3] The upper and lower respiratory tract appears to have its own set of microbiota.[101] Pulmonary bacterial microbiota belong to 9 major bacterial genera: Prevotella, Sphingomonas, Pseudomonas, Acinetobacter, Fusobacterium, Megasphaera, Veillonella, Staphylococcus, and Streptococcus. Some of the bacteria considered "normal biota" in the respiratory tract can cause serious disease especially in immunocompromised individuals; these include Streptococcus pyogenes, Haemophilus influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Staphylococcus aureus.[citation needed] Fungal genera that compose the pulmonary mycobiome include Candida, Malassezia, Neosartorya, Saccharomyces, and Aspergillus, among others.[49]
Unusual distributions of bacterial and fungal genera in the respiratory tract is observed in people with cystic fibrosis.[49][102] Their bacterial flora often contains antibiotic-resistant and slow-growing bacteria, and the frequency of these pathogens changes in relation to age.[102]
Biliary tract
[edit]Traditionally the biliary tract has been considered to be normally sterile, and the presence of microorganisms in bile is a marker of pathological process. This assumption was confirmed by failure in allocation of bacterial strains from the normal bile duct. Papers began emerging in 2013 showing that the normal biliary microbiota is a separate functional layer which protects a biliary tract from colonization by exogenous microorganisms.[103]
Disease and death
[edit]Human bodies rely on the innumerable bacterial genes as the source of essential nutrients.[104] Both metagenomic and epidemiological studies indicate vital roles for the human microbiome in preventing a wide range of diseases, from type 2 diabetes and obesity to inflammatory bowel disease, Parkinson's disease, and even mental health conditions like depression.[105] A symbiotic relationship between the gut microbiota and different bacteria may influence an individual's immune response.[106] Metabolites generated by gut microbes appear to be causative factors in type 2 diabetes.[107] Although in its infancy, microbiome-based treatment is also showing promise, most notably for treating drug-resistant C. difficile Archived 7 December 2019 at the Wayback Machine infection[108] and in diabetes treatment.[109]
Clostridioides difficile infection
[edit]An overwhelming presence of the bacteria, C. difficile, leads to an infection of the gastrointestinal tract, normally associated to dysbiosis with the microbiota believed to have been caused by the administration of antibiotics. Use of antibiotics eradicates the beneficial gut flora within the gastrointestinal tract, which normally prevents pathogenic bacteria from establishing dominance.[110] Traditional treatment for C. difficile infections includes an additional regime of antibiotics, however, efficacy rates average between 20 and 30%.[111] Recognizing the importance of healthy gut bacteria, researchers turned to a procedure known as fecal microbiota transplant (FMT), where patients experiencing gastrointestinal diseases, such as C. difficile infection (CDI), receive fecal content from a healthy individual in hopes of restoring a normal functioning intestinal microbiota.[112] Fecal microbiota transplant is approximately 85–90% effective in people with CDI for whom antibiotics have not worked or in whom the disease recurs following antibiotics.[113][114] Most people with CDI recover with one FMT treatment.[115][110][116]
Cancer
[edit]Although cancer is generally a disease of host genetics and environmental factors, microorganisms are implicated in some 20% of human cancers.[117] Particularly for potential factors in colon cancer, bacterial density is one million times higher than in the small intestine, and approximately 12-fold more cancers occur in the colon compared to the small intestine, possibly establishing a pathogenic role for microbiota in colon and rectal cancers.[118] Microbial density may be used as a prognostic tool in assessment of colorectal cancers.[118]
The microbiota may affect carcinogenesis in three broad ways: (i) altering the balance of tumor cell proliferation and death, (ii) regulating immune system function, and (iii) influencing metabolism of host-produced factors, foods and pharmaceuticals.[117] Tumors arising at boundary surfaces, such as the skin, oropharynx and respiratory, digestive and urogenital tracts, harbor a microbiota. Substantial microbe presence at a tumor site does not establish association or causal links. Instead, microbes may find tumor oxygen tension or nutrient profile supportive. Decreased populations of specific microbes or induced oxidative stress may also increase risks.[117][118] Of the around 1030 microbes on earth, ten are designated by the International Agency for Research on Cancer as human carcinogens.[117] Microbes may secrete proteins or other factors directly drive cell proliferation in the host, or may up- or down-regulate the host immune system including driving acute or chronic inflammation in ways that contribute to carcinogenesis.[117]
Concerning the relationship of immune function and development of inflammation, mucosal surface barriers are subject to environmental risks and must rapidly repair to maintain homeostasis. Compromised host or microbiota resiliency also reduce resistance to malignancy, possibly inducing inflammation and cancer. Once barriers are breached, microbes can elicit proinflammatory or immunosuppressive programs through various pathways.[117] For example, cancer-associated microbes appear to activate NF-κΒ signaling within the tumor microenvironment. Other pattern recognition receptors, such as nucleotide-binding oligomerization domain–like receptor (NLR) family members NOD-2, NLRP3, NLRP6 and NLRP12, may play a role in mediating colorectal cancer.[117] Likewise Helicobacter pylori appears to increase the risk of gastric cancer, due to its driving a chronic inflammatory response in the stomach.[117][118]
Inflammatory bowel disease
[edit]Inflammatory bowel disease consists of two different diseases: ulcerative colitis and Crohn's disease and both of these diseases present with disruptions in the gut microbiota (also known as dysbiosis). This dysbiosis presents itself in the form of decreased microbial diversity in the gut,[119][120] and is correlated to defects in host genes that changes the innate immune response in individuals.[119]
Human immunodeficiency virus
[edit]The HIV disease progression influences the composition and function of the gut microbiota, with notable differences between HIV-negative, HIV-positive, and post-ART HIV-positive populations.[citation needed] HIV decreases the integrity of the gut epithelial barrier function by affecting tight junctions. This breakdown allows for translocation across the gut epithelium, which is thought to contribute to increases in inflammation seen in people with HIV.[121]
Vaginal microbiota plays a role in the infectivity of HIV, with an increased risk of infection and transmission when the woman has bacterial vaginosis, a condition characterized by an abnormal balance of vaginal bacteria.[122] The enhanced infectivity is seen with the increase in pro-inflammatory cytokines and CCR5 + CD4+ cells in the vagina. However, a decrease in infectivity is seen with increased levels of vaginal Lactobacillus, which promotes an anti-inflammatory condition.[121]
Gut microbiome of centenarians
[edit]Humans who are 100 years old or older, called centenarians, have a distinct gut microbiome. This microbiome is characteristically enriched in microorganisms that are able to synthesize novel secondary bile acids.[123] These secondary bile acids include various isoforms of lithocholic acid that may contribute to healthy aging.[123]
Death
[edit]With death, the microbiome of the living body collapses and a different composition of microorganisms named necrobiome establishes itself as an important active constituent of the complex physical decomposition process. Its predictable changes over time are thought to be useful to help determine the time of death.[124][125]
Environmental health
[edit]Studies in 2009 questioned whether the decline in biota (including microfauna) as a result of human intervention might impede human health, hospital safety procedures, food product design, and treatments of disease.[126]
Changes, modulation and transmission
[edit]
Hygiene,[128] probiotics,[127] prebiotics,[129] synbiotics,[130] light therapy,[131] microbiota transplants (fecal[132] or skin[133]), antibiotics,[134] exercise,[135][136] diet,[137] breastfeeding,[138] aging[139] can change the human microbiome across various anatomical systems or regions such as skin and gut.
Person-to-person transmission
[edit]The human microbiome is transmitted between a mother and her children, as well as between people living in the same household.[140][141]
Research
[edit]Migration
[edit]Primary research indicates that immediate changes in the microbiota may occur when a person migrates from one country to another, such as when Thai immigrants settled in the United States[142] or when Latin Americans immigrated into the United States.[143] Losses of microbiota diversity were greater in obese individuals and children of immigrants.[142][143]
Cellulose digestion
[edit]A 2024 study suggests that gut microbiota capable of digesting cellulose can be found in the human microbiome, and they are less abundant in people living in industrialized societies.[144][145]
See also
[edit]Bibliography
[edit]- Ed Yong. I Contain Multitudes: The Microbes Within Us and a Grander View of Life. 368 pages, Published 9 August 2016 by Ecco, ISBN 0062368591.
References
[edit]- ^ a b Marchesi JR, Ravel J (2015). "The vocabulary of microbiome research: a proposal". Microbiome. 3: 31. doi:10.1186/s40168-015-0094-5. PMC 4520061. PMID 26229597.
Microbiome
This term refers to the entire habitat, including the microorganisms (bacteria, archaea, lower and higher eurkaryotes, and viruses), their genomes (i.e., genes), and the surrounding environmental conditions. This definition is based on that of "biome," the biotic and abiotic factors of given environments. Others in the field limit the definition of microbiome to the collection of genes and genomes of members of a microbiota. It is argued that this is the definition of metagenome, which combined with the environment constitutes the microbiome. - ^ Mousa, Walaa K.; Chehadeh, Fadia; Husband, Shannon (2022). "Recent Advances in Understanding the Structure and Function of the Human Microbiome". Frontiers in Microbiology. 13. doi:10.3389/fmicb.2022.825338. PMC 8851206. PMID 35185849.
- ^ a b c d e f g h i j k Sherwood L, Willey J, Woolverton C (2013). Prescott's Microbiology (9th ed.). New York: McGraw Hill. pp. 713–721. ISBN 9780073402406. OCLC 886600661.
- ^ Sender R, Fuchs S, Milo R (January 2016). "Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans". Cell. 164 (3): 337–40. doi:10.1016/j.cell.2016.01.013. PMID 26824647.
- ^ a b c d e f Quigley EM (September 2013). "Gut bacteria in health and disease". Gastroenterology & Hepatology. 9 (9): 560–9. PMC 3983973. PMID 24729765.
- ^ Falony G, Vieira-Silva S, Raes J (2015). "Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine". Annual Review of Microbiology. 69: 305–21. doi:10.1146/annurev-micro-091014-104422. PMID 26274026.
we review literature on trimethylamine (TMA), a microbiota-generated metabolite linked to atherosclerosis development.
- ^ Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF (November 2014). "Archaea and the human gut: new beginning of an old story". World Journal of Gastroenterology. 20 (43): 16062–78. doi:10.3748/wjg.v20.i43.16062. PMC 4239492. PMID 25473158.
Trimethylamine is exclusively a microbiota-derived product of nutrients (lecithin, choline, TMAO, L-carnitine) from normal diet, from which seems originate two diseases, trimethylaminuria (or Fish-Odor Syndrome) and cardiovascular disease through the proatherogenic property of its oxidized liver-derived form.
- ^ Smith A, et al. (2019). "The role of the microbiota in the development of allergies and asthma." Current Allergy and Asthma Reports, 19(8), 38.
- ^ Jackson KD, et al. (2016). "Gut microbiota associations with common diseases and prescription medications in a population-based cohort." Nature Communications, 7, 11622.
- ^ Yi DY, Kim SY (September 2021). "Human Breast Milk Composition and Function in Human Health: From Nutritional Components to Microbiome and MicroRNAs". Nutrients. 13 (9): 3094. doi:10.3390/nu13093094. PMC 8471419. PMID 34578971.
- ^ Yatsunenko T, et al. (2012). "Human gut microbiome viewed across age and geography." Nature, 486(7402), 222-227.
- ^ Sjögren YM, et al. (2009). "Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses." Clinical and Experimental Allergy, 39(12), 1842-1851.
- ^ a b "NIH Human Microbiome Project defines normal bacterial makeup of the body". NIH News. 13 June 2012.
- ^ Sender, Ron; Fuchs, Shai; Milo, Ron (19 August 2016). "Revised Estimates for the Number of Human and Bacteria Cells in the Body". PLOS Biology. 14 (8): e1002533. doi:10.1371/journal.pbio.1002533. ISSN 1545-7885. PMC 4991899. PMID 27541692.
- ^ Lacy, Brian E.; Spiegel, Brennan (July 2019). "Introduction to the Gut Microbiome Special Issue". American Journal of Gastroenterology. 114 (7): 1013. doi:10.14309/ajg.0000000000000303. ISSN 0002-9270. PMID 31205134.
- ^ Sung, Jaeyun; Rajendraprasad, Sanu S.; Philbrick, Kemuel L.; Bauer, Brent A.; Gajic, Ognjen; Shah, Aditya; Laudanski, Krzysztof; Bakken, Johan S.; Skalski, Joseph; Karnatovskaia, Lioudmila V. (February 2024). "The human gut microbiome in critical illness: disruptions, consequences, and therapeutic frontiers". Journal of Critical Care. 79: 154436. doi:10.1016/j.jcrc.2023.154436. ISSN 1557-8615. PMC 11034825. PMID 37769422.
- ^ Abbott, Alison (8 January 2016). "Scientists bust myth that our bodies have more bacteria than human cells". Nature. doi:10.1038/nature.2016.19136. ISSN 1476-4687. S2CID 190879263.
- ^ Gilbert, Jack A.; Blaser, Martin J.; Caporaso, J. Gregory; Jansson, Janet K.; Lynch, Susan V.; Knight, Rob (10 April 2018). "Current understanding of the human microbiome". Nature Medicine. 24 (4): 392–400. doi:10.1038/nm.4517. ISSN 1546-170X. PMC 7043356. PMID 29634682.
- ^ Wischmeyer, Paul E.; McDonald, Daniel; Knight, Rob (August 2016). "Role of the microbiome, probiotics, and 'dysbiosis therapy' in critical illness". Current Opinion in Critical Care. 22 (4): 347–353. doi:10.1097/MCC.0000000000000321. ISSN 1531-7072. PMC 5065053. PMID 27327243.
- ^ Qin, Junjie; Li, Ruiqiang; Raes, Jeroen; Arumugam, Manimozhiyan; Burgdorf, Kristoffer Solvsten; Manichanh, Chaysavanh; Nielsen, Trine; Pons, Nicolas; Levenez, Florence; Yamada, Takuji; Mende, Daniel R.; Li, Junhua; Xu, Junming; Li, Shaochuan; Li, Dongfang (4 March 2010). "A human gut microbial gene catalogue established by metagenomic sequencing". Nature. 464 (7285): 59–65. Bibcode:2010Natur.464...59.. doi:10.1038/nature08821. ISSN 1476-4687. PMC 3779803. PMID 20203603.
- ^ a b c Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. (December 2009). "The NIH Human Microbiome Project". Genome Research. 19 (12): 2317–23. doi:10.1101/gr.096651.109. PMC 2792171. PMID 19819907.
- ^ Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R (December 2011). "Experimental and analytical tools for studying the human microbiome". Nature Reviews. Genetics. 13 (1): 47–58. doi:10.1038/nrg3129. PMC 5119550. PMID 22179717.
- ^ Vestheim H, Jarman SN (July 2008). "Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs". Frontiers in Zoology. 5: 12. doi:10.1186/1742-9994-5-12. PMC 2517594. PMID 18638418.
- ^ Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. (October 2009). "Towards the human intestinal microbiota phylogenetic core". Environmental Microbiology. 11 (10): 2574–84. Bibcode:2009EnvMi..11.2574T. doi:10.1111/j.1462-2920.2009.01982.x. PMID 19601958.
- ^ Hamady M, Knight R (July 2009). "Microbial community profiling for human microbiome projects: Tools, techniques, and challenges". Genome Research. 19 (7): 1141–52. doi:10.1101/gr.085464.108. PMC 3776646. PMID 19383763.
- ^ Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, et al. (Human Microbiome Project Consortium) (June 2012). "A framework for human microbiome research". Nature. 486 (7402): 215–21. Bibcode:2012Natur.486..215T. doi:10.1038/nature11209. PMC 3377744. PMID 22699610.
- ^ The Human Microbiome Project Consortium (June 2012). "Structure, function and diversity of the healthy human microbiome". Nature. 486 (7402): 207–14. Bibcode:2012Natur.486..207T. doi:10.1038/nature11234. PMC 3564958. PMID 22699609.
- ^ "NIH Human Microbiome Project defines normal bacterial makeup of the body". National Institutes of Health (NIH). 31 August 2015. Retrieved 7 July 2023.
- ^ Quince C, Walker AW, Simpson JT, Loman NJ, Segata N (September 2017). "Shotgun metagenomics, from sampling to analysis" (PDF). Nature Biotechnology. 35 (9): 833–844. doi:10.1038/nbt.3935. hdl:2164/10167. PMID 28898207. S2CID 19041044.
- ^ Claesson MJ, Clooney AG, O'Toole PW (October 2017). "A clinician's guide to microbiome analysis". Nature Reviews. Gastroenterology & Hepatology. 14 (10): 585–595. doi:10.1038/nrgastro.2017.97. PMID 28790452. S2CID 24644894.
- ^ a b c Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. (July 2018). "Best practices for analysing microbiomes". Nature Reviews. Microbiology. 16 (7): 410–422. doi:10.1038/s41579-018-0029-9. PMID 29795328. S2CID 43936002.
- ^ a b Washburne AD, Morton JT, Sanders J, McDonald D, Zhu Q, Oliverio AM, Knight R (June 2018). "Methods for phylogenetic analysis of microbiome data". Nature Microbiology. 3 (6): 652–661. doi:10.1038/s41564-018-0156-0. PMID 29795540. S2CID 43962376.
- ^ Berg, Gabriele; et al. (2020). "Microbiome definition re-visited: Old concepts and new challenges". Microbiome. 8 (1): 103. doi:10.1186/s40168-020-00875-0. PMC 7329523. PMID 32605663.
- ^ Faust, Karoline; Raes, Jeroen (2016). "Rules of the game for microbiota". Nature. 534 (7606): 182–183. doi:10.1038/534182a. PMID 27279206. S2CID 205089271.
- ^ Bashan, Amir; Gibson, Travis E.; Friedman, Jonathan; Carey, Vincent J.; Weiss, Scott T.; Hohmann, Elizabeth L.; Liu, Yang-Yu (2016). "Universality of human microbial dynamics". Nature. 534 (7606): 259–262. Bibcode:2016Natur.534..259B. doi:10.1038/nature18301. PMC 4902290. PMID 27279224.
- ^ Liu, Yang-Yu (15 February 2023). "Controlling the human microbiome" (PDF). Cell Systems. 14 (2): 135–159. doi:10.1016/j.cels.2022.12.010. PMC 9942095. PMID 36796332.
- ^ Kennedy, Katherine M.; de Goffau, Marcus C.; Perez-Muñoz, Maria Elisa; Arrieta, Marie-Claire; Bäckhed, Fredrik; Bork, Peer; Braun, Thorsten; Bushman, Frederic D.; Dore, Joel; de Vos, Willem M.; Earl, Ashlee M.; Eisen, Jonathan A.; Elovitz, Michal A.; Ganal-Vonarburg, Stephanie C.; Gänzle, Michael G. (January 2023). "Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies". Nature. 613 (7945): 639–649. Bibcode:2023Natur.613..639K. doi:10.1038/s41586-022-05546-8. hdl:2164/21298. ISSN 1476-4687. PMC 11333990. PMID 36697862. S2CID 256268291.
- ^ PLoS Human Microbiome Project Collection Manuscript Summaries Archived 4 March 2014 at the Wayback Machine 13 June 2012
- ^ "Consortium of Scientists Map the Human Body's Bacterial Ecosystem". ucsf.edu. 13 June 2012.
- ^ Sommer F, Bäckhed F (April 2013). "The gut microbiota--masters of host development and physiology". Nature Reviews. Microbiology. 11 (4): 227–38. doi:10.1038/nrmicro2974. PMID 23435359. S2CID 22798964.
- ^ Gajer, Pawel; Brotman, Rebecca M.; Bai, Guoyun; Sakamoto, Joyce; Schütte, Ursel M. E.; Zhong, Xue; Koenig, Sara S. K.; Fu, Li; Ma, Zhanshan (Sam); Zhou, Xia; Abdo, Zaid; Forney, Larry J.; Ravel, Jacques (2 May 2012). "Temporal Dynamics of the Human Vaginal Microbiota". Science Translational Medicine. 4 (132): 132ra52. doi:10.1126/scitranslmed.3003605. ISSN 1946-6234. PMC 3722878. PMID 22553250.
- ^ Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. (June 2005). "Diversity of the human intestinal microbial flora". Science. 308 (5728): 1635–8. Bibcode:2005Sci...308.1635E. doi:10.1126/science.1110591. PMC 1395357. PMID 15831718.
- ^ Duncan SH, Louis P, Flint HJ (April 2007). "Cultivable bacterial diversity from the human colon". Letters in Applied Microbiology. 44 (4): 343–50. doi:10.1111/j.1472-765X.2007.02129.x. PMID 17397470. S2CID 43706882.
- ^ Florin TH, Zhu G, Kirk KM, Martin NG (October 2000). "Shared and unique environmental factors determine the ecology of methanogens in humans and rats". The American Journal of Gastroenterology. 95 (10): 2872–9. CiteSeerX 10.1.1.606.4187. doi:10.1111/j.1572-0241.2000.02319.x. PMID 11051362. S2CID 1087298.
- ^ Eckburg PB, Lepp PW, Relman DA (February 2003). "Archaea and their potential role in human disease". Infection and Immunity. 71 (2): 591–6. doi:10.1128/IAI.71.2.591-596.2003. PMC 145348. PMID 12540534.
- ^ Cavicchioli R, Curmi PM, Saunders N, Thomas T (November 2003). "Pathogenic archaea: do they exist?". BioEssays. 25 (11): 1119–28. doi:10.1002/bies.10354. PMID 14579252.
- ^ Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA (April 2004). "Methanogenic Archaea and human periodontal disease". Proceedings of the National Academy of Sciences of the United States of America. 101 (16): 6176–81. Bibcode:2004PNAS..101.6176L. doi:10.1073/pnas.0308766101. PMC 395942. PMID 15067114.
- ^ Takakura, Will; Pimentel, Mark (10 July 2020). "Small Intestinal Bacterial Overgrowth and Irritable Bowel Syndrome – An Update". Frontiers in Psychiatry. 11: 664. doi:10.3389/fpsyt.2020.00664. ISSN 1664-0640. PMC 7366247. PMID 32754068.
- ^ a b c d e f g h i j k Cui L, Morris A, Ghedin E (July 2013). "The human mycobiome in health and disease". Genome Medicine. 5 (7): 63. doi:10.1186/gm467. PMC 3978422. PMID 23899327. Figure 2: Distribution of fungal genera in different body sites
- ^ a b Martins N, Ferreira IC, Barros L, Silva S, Henriques M (June 2014). "Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment" (PDF). Mycopathologia. 177 (5–6): 223–40. doi:10.1007/s11046-014-9749-1. hdl:10198/10147. PMID 24789109. S2CID 795450.
Candida species and other microorganisms are involved in this complicated fungal infection, but Candida albicans continues to be the most prevalent. In the past two decades, it has been observed an abnormal overgrowth in the gastrointestinal, urinary and respiratory tracts, not only in immunocompromised patients, but also related to nosocomial infections and even in healthy individuals. There is a widely variety of causal factors that contribute to yeast infection which means that candidiasis is a good example of a multifactorial syndrome.
- ^ a b c d e Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH (April 2014). "Review article: fungal microbiota and digestive diseases". Alimentary Pharmacology & Therapeutics. 39 (8): 751–66. doi:10.1111/apt.12665. PMID 24612332. S2CID 22101484.
In addition, GI fungal infection is reported even among those patients with normal immune status. Digestive system-related fungal infections may be induced by both commensal opportunistic fungi and exogenous pathogenic fungi. ... Candida sp. is also the most frequently identified species among patients with gastric IFI. ... It was once believed that gastric acid could kill microbes entering the stomach and that the unique ecological environment of the stomach was not suitable for microbial colonisation or infection. However, several studies using culture-independent methods confirmed that large numbers of acid-resistant bacteria belonging to eight phyla and up to 120 species exist in the stomach, such as Streptococcus sp., Neisseria sp. and Lactobacillus sp. etc.26, 27 Furthermore, Candida albicans can grow well in highly acidic environments,28 and some genotypes may increase the severity of gastric mucosal lesions.29
- ^ a b c Erdogan A, Rao SS (April 2015). "Small intestinal fungal overgrowth". Current Gastroenterology Reports. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900. S2CID 3098136.
Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. Two recent studies showed that 26 % (24/94) and 25.3 % (38/150) of a series of patients with unexplained GI symptoms had SIFO. The most common symptoms observed in these patients were belching, bloating, indigestion, nausea, diarrhea, and gas. ... Fungal-bacterial interaction may act in different ways and may either be synergistic or antagonistic or symbiotic [29]. Some bacteria such as Lactobacillus species can interact and inhibit both the virulence and growth of Candida species in the gut by producing hydrogen peroxide [30]. Any damage to the mucosal barrier or disruption of GI microbiota with chemotherapy or antibiotic use, inflammatory processes, activation of immune molecules and disruption of epithelial repair may all cause fungal overgrowth [27].
- ^ Marcon MJ, Powell DA (April 1992). "Human infections due to Malassezia spp". Clinical Microbiology Reviews. 5 (2): 101–19. doi:10.1128/CMR.5.2.101. PMC 358230. PMID 1576583.
- ^ Roth RR, James WD (1988). "Microbial ecology of the skin". Annual Review of Microbiology. 42 (1): 441–64. doi:10.1146/annurev.mi.42.100188.002301. PMID 3144238.
- ^ Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. (October 2015). "The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome". mBio. 6 (5): e01578-15. doi:10.1128/mBio.01578-15. PMC 4620475. PMID 26489866.
- ^ Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. (October 2011). "The human gut virome: inter-individual variation and dynamic response to diet". Genome Research. 21 (10): 1616–25. doi:10.1101/gr.122705.111. PMC 3202279. PMID 21880779.
- ^ Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. (January 2015). "Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients". American Journal of Transplantation. 15 (1): 200–9. doi:10.1111/ajt.13031. PMC 4276431. PMID 25403800.
- ^ Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT (September 2014). "Human oral viruses are personal, persistent and gender-consistent". The ISME Journal. 8 (9): 1753–67. Bibcode:2014ISMEJ...8.1753A. doi:10.1038/ismej.2014.31. PMC 4139723. PMID 24646696.
- ^ Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT (May 2014). "Altered oral viral ecology in association with periodontal disease". mBio. 5 (3): e01133-14. doi:10.1128/mBio.01133-14. PMC 4030452. PMID 24846382.
- ^ Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. (March 2016). "Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome". Cell Host & Microbe. 19 (3): 311–22. doi:10.1016/j.chom.2016.02.011. PMC 4821831. PMID 26962942.
- ^ Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. (January 2015). "Disease-specific alterations in the enteric virome in inflammatory bowel disease". Cell. 160 (3): 447–60. doi:10.1016/j.cell.2015.01.002. PMC 4312520. PMID 25619688.
- ^ Koumoundouros, Tessa (29 January 2024). "'Obelisks': Entirely New Class of Life Has Been Found in The Human Digestive System". ScienceAlert. Archived from the original on 29 January 2024. Retrieved 29 January 2024.
- ^ Zheludev, Ivan N.; et al. (21 January 2024). "Viroid-like colonists of human microbiomes". bioRxiv: 2024.01.20.576352. doi:10.1101/2024.01.20.576352. PMC 10827157. PMID 38293115. Archived from the original on 29 January 2024. Retrieved 29 January 2024.
- ^ Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. (NISC Comparative Sequencing Program) (May 2009). "Topographical and temporal diversity of the human skin microbiome". Science. 324 (5931): 1190–2. Bibcode:2009Sci...324.1190G. doi:10.1126/science.1171700. PMC 2805064. PMID 19478181.
- ^ a b "The Normal Bacterial Flora of Humans". textbookofbacteriology.net.
- ^ a b c d e f g h i Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492. PMID 27102537.
Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease - ^ Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (March 2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–3703. Bibcode:2009PNAS..106.3698W. doi:10.1073/pnas.0812874106. PMC 2656143. PMID 19234110.
Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes.
IPA metabolism diagram - ^ "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 June 2018.
- ^ Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (July 1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–21942. doi:10.1074/jbc.274.31.21937. PMID 10419516. S2CID 6630247.
[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
- ^ Yang I, Corwin EJ, Brennan PA, Jordan S, Murphy JR, Dunlop A (2016). "The Infant Microbiome: Implications for Infant Health and Neurocognitive Development". Nursing Research. 65 (1): 76–88. doi:10.1097/NNR.0000000000000133. PMC 4681407. PMID 26657483.
- ^ Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG (February 2015). "The infant microbiome development: mom matters". Trends in Molecular Medicine. 21 (2): 109–17. doi:10.1016/j.molmed.2014.12.002. PMC 4464665. PMID 25578246.
- ^ Wall R, Ross RP, Ryan CA, Hussey S, Murphy B, Fitzgerald GF, Stanton C (4 March 2009). "Role of gut microbiota in early infant development". Clinical Medicine. Pediatrics. 3: 45–54. doi:10.4137/cmped.s2008. PMC 3676293. PMID 23818794.
- ^ a b c Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG (August 2014). "Minireview: Gut microbiota: the neglected endocrine organ". Molecular Endocrinology. 28 (8): 1221–38. doi:10.1210/me.2014-1108. PMC 5414803. PMID 24892638.
- ^ a b Shen S, Wong CH (April 2016). "Bugging inflammation: role of the gut microbiota". Clinical & Translational Immunology. 5 (4): e72. doi:10.1038/cti.2016.12. PMC 4855262. PMID 27195115.
- ^ Wang H, Lee IS, Braun C, Enck P (October 2016). "Effect of Probiotics on Central Nervous System Functions in Animals and Humans: A Systematic Review". Journal of Neurogastroenterology and Motility. 22 (4): 589–605. doi:10.5056/jnm16018. PMC 5056568. PMID 27413138.
These probiotics showed efficacy in improving psychiatric disorder-related behaviors including anxiety, depression, autism spectrum disorder (ASD), obsessive-compulsive disorder, and memory abilities, including spatial and non-spatial memory. Because many of the basic science studies showed some efficacy of probiotics on central nervous system function, this background may guide and promote further preclinical and clinical studies. ... According to the qualitative analyses of current studies, we can provisionally draw the conclusion that B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei were most effective in improving CNS function, including psychiatric disease-associated functions (anxiety, depression, mood, stress response) and memory abilities
- ^ a b Drake MJ, Morris N, Apostolidis A, Rahnama'i MS, Marchesi JR (April 2017). "The urinary microbiome and its contribution to lower urinary tract symptoms; ICI-RS 2015". Neurourology and Urodynamics. 36 (4): 850–853. doi:10.1002/nau.23006. hdl:1983/3b024f95-9f86-406a-9be3-ce35984b8de1. PMID 28444712. S2CID 27636043.
- ^ a b Aragón IM, Herrera-Imbroda B, Queipo-Ortuño MI, Castillo E, Del Moral JS, Gómez-Millán J, et al. (January 2018). "The Urinary Tract Microbiome in Health and Disease". European Urology Focus. 4 (1): 128–138. doi:10.1016/j.euf.2016.11.001. PMID 28753805.
- ^ a b Schmiemann G, Kniehl E, Gebhardt K, Matejczyk MM, Hummers-Pradier E (May 2010). "The diagnosis of urinary tract infection: a systematic review". Deutsches Ärzteblatt International. 107 (21): 361–7. doi:10.3238/arztebl.2010.0361. PMC 2883276. PMID 20539810.
- ^ Al, Kait F.; Denstedt, John D.; Daisley, Brendan A.; Bjazevic, Jennifer; Welk, Blayne K.; Pautler, Stephen E.; Gloor, Gregory B.; Reid, Gregor; Razvi, Hassan; Burton, Jeremy P. (September 2020). "Ureteral Stent Microbiota Is Associated with Patient Comorbidities but Not Antibiotic Exposure". Cell Reports Medicine. 1 (6): 100094. doi:10.1016/j.xcrm.2020.100094. PMC 7659606. PMID 33205072.
- ^ Wolfe AJ, Brubaker L (February 2019). "Urobiome updates: advances in urinary microbiome research". Nature Reviews. Urology. 16 (2): 73–74. doi:10.1038/s41585-018-0127-5. PMC 6628711. PMID 30510275.
- ^ a b Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S (2015). "Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health". Frontiers in Physiology. 6: 81. doi:10.3389/fphys.2015.00081. PMC 4373506. PMID 25859220.
- ^ a b c d Witkin SS, Linhares IM, Giraldo P (June 2007). "Bacterial flora of the female genital tract: function and immune regulation". Best Practice & Research. Clinical Obstetrics & Gynaecology. 21 (3): 347–54. doi:10.1016/j.bpobgyn.2006.12.004. PMID 17215167.
- ^ Todar K (2012). "The Normal Bacterial Flora of Humans". Todar's Online Textbook of Bacteriology. Madison, WI: Kenneth Todar. Retrieved 6 April 2012.
- ^ Onderdonk AB, Zamarchi GR, Walsh JA, Mellor RD, Muñoz A, Kass EH (February 1986). "Methods for quantitative and qualitative evaluation of vaginal microflora during menstruation". Applied and Environmental Microbiology. 51 (2): 333–9. Bibcode:1986ApEnM..51..333O. doi:10.1128/AEM.51.2.333-339.1986. PMC 238869. PMID 3954346.
- ^ Antonio MA, Hawes SE, Hillier SL (December 1999). "The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species". The Journal of Infectious Diseases. 180 (6): 1950–6. doi:10.1086/315109. PMID 10558952.
- ^ Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, et al. (March 2020). "Cervicovaginal microbiome and natural history of HPV in a longitudinal study". PLOS Pathogens. 16 (3): e1008376. doi:10.1371/journal.ppat.1008376. PMC 7098574. PMID 32214382.
- ^ Fox C, Eichelberger K (December 2015). "Maternal microbiome and pregnancy outcomes". Fertility and Sterility. 104 (6): 1358–63. doi:10.1016/j.fertnstert.2015.09.037. PMID 26493119.
- ^ Wassenaar TM, Panigrahi P (December 2014). "Is a foetus developing in a sterile environment?". Letters in Applied Microbiology. 59 (6): 572–9. doi:10.1111/lam.12334. PMID 25273890. S2CID 206169539.
- ^ Schwiertz A (2016). Microbiota of the human body : implications in health and disease. Switzerland: Springer. p. 1. ISBN 978-3-319-31248-4.
- ^ Tamburini S, Shen N, Wu HC, Clemente JC (July 2016). "The microbiome in early life: implications for health outcomes". Nature Medicine. 22 (7): 713–22. doi:10.1038/nm.4142. PMID 27387886. S2CID 2462790.
- ^ de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. (August 2019). "Human placenta has no microbiome but can contain potential pathogens". Nature. 572 (7769): 329–334. Bibcode:2019Natur.572..329D. doi:10.1038/s41586-019-1451-5. PMC 6697540. PMID 31367035.
- ^ Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS (February 2019). "Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations". Trends in Microbiology. 27 (2): 105–117. doi:10.1016/j.tim.2018.11.003. hdl:2440/122027. PMID 30497919. S2CID 54166123.
- ^ Franasiak JM, Scott RT (December 2015). "Reproductive tract microbiome in assisted reproductive technologies". Fertility and Sterility. 104 (6): 1364–71. doi:10.1016/j.fertnstert.2015.10.012. PMID 26597628.
- ^ Sutter VL (1984). "Anaerobes as normal oral flora". Reviews of Infectious Diseases. 6 (Suppl 1): S62-566. doi:10.1093/clinids/6.Supplement_1.S62. PMID 6372039.
- ^ a b c d e Kumar PS (December 2013). "Oral microbiota and systemic disease". Anaerobe. 24: 90–3. doi:10.1016/j.anaerobe.2013.09.010. PMID 24128801. S2CID 40735283.
- ^ Arweiler NB, Netuschil L (May 2016). "The Oral Microbiota". In Schwiertz A (ed.). Microbiota of the Human Body. Advances in Experimental Medicine and Biology. Vol. 902. Springer, Cham. pp. 45–60. doi:10.1007/978-3-319-31248-4_4. ISBN 978-3-319-31248-4. PMID 27161350.
- ^ a b c d Avila M, Ojcius DM, Yilmaz O (August 2009). "The oral microbiota: living with a permanent guest". DNA and Cell Biology. 28 (8): 405–11. doi:10.1089/dna.2009.0874. PMC 2768665. PMID 19485767.
- ^ a b Rogers AH, ed. (2008). Molecular Oral Microbiology. Caister Academic Press. ISBN 978-1-904455-24-0.
- ^ a b c d e Zarco MF, Vess TJ, Ginsburg GS (March 2012). "The oral microbiome in health and disease and the potential impact on personalized dental medicine". Oral Diseases. 18 (2): 109–20. doi:10.1111/j.1601-0825.2011.01851.x. PMID 21902769. S2CID 24411104.
- ^ Rhoades NS, Pinski AN, Monsibais AN, Jankeel A, Doratt BM, Cinco IR, et al. (August 2021). "Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose". Cell Reports. 36 (9): 109637. doi:10.1016/j.celrep.2021.109637. ISSN 2211-1247. PMC 8361213. PMID 34433082.
- ^ Wing Ho Man; Wouter A A de Steenhuijsen Piters; Debby Bogaert (2017). "The microbiota of the respiratory tract: gatekeeper to respiratory health". Nature Reviews Microbiology. 15 (5) (published 20 March 2017): 259–270. doi:10.1038/NRMICRO.2017.14. hdl:20.500.11820/f1137874-9c51-401b-bca4-e2a5da3e219b. ISSN 1740-1534. PMC 7097736. PMID 28316330.Wikidata: Q34553608
- ^ a b Beringer PM, Appleman MD (November 2000). "Unusual respiratory bacterial flora in cystic fibrosis: microbiologic and clinical features" (PDF). Current Opinion in Pulmonary Medicine. 6 (6): 545–50. doi:10.1097/00063198-200011000-00015. PMID 11100967. S2CID 845977. Archived from the original (PDF) on 16 October 2013.
- ^ Verdier J, Luedde T, Sellge G (June 2015). "Biliary Mucosal Barrier and Microbiome". Viszeralmedizin. 31 (3): 156–61. doi:10.1159/000431071 (inactive 21 November 2024). PMC 4569210. PMID 26468308.
{{cite journal}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Yu B, Yu B, Yu L (June 2020). "Commentary: Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome". Indian Journal of Microbiology. 60 (2): 259–261. doi:10.1007/s12088-020-00863-w. PMC 7105528. PMID 32255860.
- ^ Copeland CS. The World Within Us: Health and the Human Microbiome. Healthcare Journal of New Orleans, Sept-Oct 2017.
- ^ Honda K, Littman DR (July 2016). "The microbiota in adaptive immune homeostasis and disease". Nature. 535 (7610): 75–84. Bibcode:2016Natur.535...75H. doi:10.1038/nature18848. PMID 27383982. S2CID 4461492.
- ^ "Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes". Metabolon. Retrieved 3 November 2022.
- ^ Liubakka A, Vaughn BP (July 2016). "Clostridium difficile Infection and Fecal Microbiota Transplant". AACN Advanced Critical Care. 27 (3): 324–337. doi:10.4037/aacnacc2016703. PMC 5666691. PMID 27959316.
- ^ Burton JH, Johnson M, Johnson J, Hsia DS, Greenway FL, Heiman ML (July 2015). "Addition of a Gastrointestinal Microbiome Modulator to Metformin Improves Metformin Tolerance and Fasting Glucose Levels". Journal of Diabetes Science and Technology. 9 (4): 808–14. doi:10.1177/1932296815577425. PMC 4525649. PMID 25802471.
- ^ a b Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, et al. (December 2011). "Treating Clostridium difficile infection with fecal microbiota transplantation". Clinical Gastroenterology and Hepatology. 9 (12): 1044–9. doi:10.1016/j.cgh.2011.08.014. PMC 3223289. PMID 21871249.
- ^ Gough E, Shaikh H, Manges AR (November 2011). "Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection". Clinical Infectious Diseases. 53 (10): 994–1002. doi:10.1093/cid/cir632. PMID 22002980.
- ^ Brown WR (August 2014). "Fecal microbiota transplantation in treating Clostridium difficile infection". Journal of Digestive Diseases. 15 (8): 405–8. doi:10.1111/1751-2980.12160. PMID 24825534. S2CID 44651256.
- ^ Burke KE, Lamont JT (August 2013). "Fecal transplantation for recurrent Clostridium difficile infection in older adults: a review". Journal of the American Geriatrics Society. 61 (8): 1394–8. doi:10.1111/jgs.12378. PMID 23869970. S2CID 34998497.
- ^ Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, et al. (May 2015). "Fecal Microbiota Transplantation for Clostridium difficile Infection: A Systematic Review". Annals of Internal Medicine. 162 (9): 630–8. doi:10.7326/m14-2693. PMID 25938992. S2CID 1307726.
- ^ Brandt LJ, Borody TJ, Campbell J (September 2011). "Endoscopic fecal microbiota transplantation: "first-line" treatment for severe clostridium difficile infection?". Journal of Clinical Gastroenterology. 45 (8): 655–7. doi:10.1097/MCG.0b013e3182257d4f. PMID 21716124. S2CID 2508836.
- ^ Kelly CR, de Leon L, Jasutkar N (February 2012). "Fecal microbiota transplantation for relapsing Clostridium difficile infection in 26 patients: methodology and results". Journal of Clinical Gastroenterology. 46 (2): 145–9. doi:10.1097/MCG.0b013e318234570b. PMID 22157239. S2CID 30849491.
- ^ a b c d e f g h Garrett WS (April 2015). "Cancer and the microbiota". Science. 348 (6230): 80–6. Bibcode:2015Sci...348...80G. doi:10.1126/science.aaa4972. PMC 5535753. PMID 25838377.
- ^ a b c d Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. (January 2016). "Gut microbiota imbalance and colorectal cancer". World Journal of Gastroenterology. 22 (2): 501–18. doi:10.3748/wjg.v22.i2.501. PMC 4716055. PMID 26811603.
- ^ a b Sartor RB, Mazmanian SK (July 2012). "Intestinal Microbes in Inflammatory Bowel Diseases". The American Journal of Gastroenterology Supplements. 1 (1): 15–21. doi:10.1038/ajgsup.2012.4.
- ^ Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I (February 2014). "Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years?". World Journal of Gastroenterology. 20 (5): 1192–210. doi:10.3748/wjg.v20.i5.1192. PMC 3921503. PMID 24574795.
- ^ a b Zilberman-Schapira G, Zmora N, Itav S, Bashiardes S, Elinav H, Elinav E (June 2016). "The gut microbiome in human immunodeficiency virus infection". BMC Medicine. 14 (1): 83. doi:10.1186/s12916-016-0625-3. PMC 4891875. PMID 27256449.
- ^ Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S (September 2013). "Vaginal microbiota and its role in HIV transmission and infection". FEMS Microbiology Reviews. 37 (5): 762–92. doi:10.1111/1574-6976.12029. PMID 23789590. S2CID 32045698.
- ^ a b Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, Skelly AN, Okamura Y, Vlamakis H, Li Y, Tanoue T, Takei H, Nittono H, Narushima S, Irie J, Itoh H, Moriya K, Sugiura Y, Suematsu M, Moritoki N, Shibata S, Littman DR, Fischbach MA, Uwamino Y, Inoue T, Honda A, Hattori M, Murai T, Xavier RJ, Hirose N, Honda K. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021 Nov;599(7885):458-464. doi: 10.1038/s41586-021-03832-5. Epub 2021 Jul 29. PMID 34325466
- ^ Intagliata C (22 December 2016). ""Necrobiome" Reveals a Corpse's Time of Death". Scientific American. Retrieved 26 March 2018.
- ^ Young E (10 December 2015). "Meet the Necrobiome: The Waves of Microbes That Will Eat Your Corpse". The Atlantic. Retrieved 26 March 2018.
- ^ Harmon K (16 December 2009). "Bugs Inside: What Happens When the Microbes That Keep Us Healthy Disappear?". Scientific American. Retrieved 27 December 2008.
- ^ a b Hitch, Thomas C. A.; Hall, Lindsay J.; Walsh, Sarah Kate; Leventhal, Gabriel E.; Slack, Emma; de Wouters, Tomas; Walter, Jens; Clavel, Thomas (June 2022). "Microbiome-based interventions to modulate gut ecology and the immune system". Mucosal Immunology. 15 (6): 1095–1113. doi:10.1038/s41385-022-00564-1. ISSN 1935-3456. PMC 9705255. PMID 36180583.
- ^ Yu, Boxuan; Yu, Bowei; Yu, Ligen (June 2020). "Commentary: Reconciling Hygiene and Cleanliness: A New Perspective from Human Microbiome". Indian Journal of Microbiology. 60 (2): 259–261. doi:10.1007/s12088-020-00863-w. ISSN 0046-8991. PMC 7105528. PMID 32255860.
- ^ Roberfroid, Marcel; Gibson, Glenn R.; Hoyles, Lesley; McCartney, Anne L.; Rastall, Robert; Rowland, Ian; Wolvers, Danielle; Watzl, Bernhard; Szajewska, Hania; Stahl, Bernd; Guarner, Francisco; Respondek, Frederique; Whelan, Kevin; Coxam, Veronique; Davicco, Marie-Jeanne (August 2010). "Prebiotic effects: metabolic and health benefits". The British Journal of Nutrition. 104 (Suppl 2): S1–63. doi:10.1017/S0007114510003363. ISSN 1475-2662. PMID 20920376.
- ^ Alli, Sauliha R.; Gorbovskaya, Ilona; Liu, Jonathan C. W.; Kolla, Nathan J.; Brown, Lisa; Müller, Daniel J. (19 April 2022). "The Gut Microbiome in Depression and Potential Benefit of Prebiotics, Probiotics and Synbiotics: A Systematic Review of Clinical Trials and Observational Studies". International Journal of Molecular Sciences. 23 (9): 4494. doi:10.3390/ijms23094494. ISSN 1422-0067. PMC 9101152. PMID 35562885.
- ^ Ellis, Jeremy R.; Powell, Eron J.; Tomasovic, Luke M.; Marcheskie, Rachel L.; Girish, Vishruth; Warman, Anmol; Sivaloganathan, Darshan (June 2024). "Changes in the Skin Microbiome Following Dermatological Procedures: A Scoping Review". Applied Microbiology. 4 (2): 972–985. doi:10.3390/applmicrobiol4020066. ISSN 2673-8007.
- ^ Zhou, Siyu; Cui, Ying; Zhang, Yun; Zhao, Tianyu; Cong, Jing (8 March 2023). "Fecal microbiota transplantation for induction of remission in Crohn's disease: a systematic review and meta-analysis". International Journal of Colorectal Disease. 38 (1): 62. doi:10.1007/s00384-023-04354-4. ISSN 1432-1262. PMID 36882658.
- ^ Callewaert, Chris; Knödlseder, Nastassia; Karoglan, Ante; Güell, Marc; Paetzold, Bernhard (1 January 2021). "Skin microbiome transplantation and manipulation: Current state of the art". Computational and Structural Biotechnology Journal. 19: 624–631. doi:10.1016/j.csbj.2021.01.001. ISSN 2001-0370. PMC 7806958. PMID 33510866.
- ^ McDonnell, Lucy; Gilkes, Alexander; Ashworth, Mark; Rowland, Victoria; Harries, Timothy Hugh; Armstrong, David; White, Patrick (2021). "Association between antibiotics and gut microbiome dysbiosis in children: systematic review and meta-analysis". Gut Microbes. 13 (1): 1–18. doi:10.1080/19490976.2020.1870402. ISSN 1949-0984. PMC 7928022. PMID 33651651.
- ^ Monda, Vincenzo; Villano, Ines; Messina, Antonietta; Valenzano, Anna; Esposito, Teresa; Moscatelli, Fiorenzo; Viggiano, Andrea; Cibelli, Giuseppe; Chieffi, Sergio; Monda, Marcellino; Messina, Giovanni (5 March 2017). "Exercise Modifies the Gut Microbiota with Positive Health Effects". Oxidative Medicine and Cellular Longevity. 2017: e3831972. doi:10.1155/2017/3831972. ISSN 1942-0900. PMC 5357536. PMID 28357027.
- ^ Mailing, Lucy J.; Allen, Jacob M.; Buford, Thomas W.; Fields, Christopher J.; Woods, Jeffrey A. (April 2019). "Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health". Exercise and Sport Sciences Reviews. 47 (2): 75–85. doi:10.1249/JES.0000000000000183. ISSN 0091-6331. PMID 30883471. S2CID 83461620.
- ^ So, Daniel; Whelan, Kevin; Rossi, Megan; Morrison, Mark; Holtmann, Gerald; Kelly, Jaimon T.; Shanahan, Erin R.; Staudacher, Heidi M.; Campbell, Katrina L. (1 June 2018). "Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis". The American Journal of Clinical Nutrition. 107 (6): 965–983. doi:10.1093/ajcn/nqy041. hdl:10536/DRO/DU:30117355. ISSN 1938-3207. PMID 29757343.
- ^ Ho, Nhan T.; Li, Fan; Lee-Sarwar, Kathleen A.; Tun, Hein M.; Brown, Bryan P.; Pannaraj, Pia S.; Bender, Jeffrey M.; Azad, Meghan B.; Thompson, Amanda L.; Weiss, Scott T.; Azcarate-Peril, M. Andrea; Litonjua, Augusto A.; Kozyrskyj, Anita L.; Jaspan, Heather B.; Aldrovandi, Grace M. (9 October 2018). "Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations". Nature Communications. 9 (1): 4169. Bibcode:2018NatCo...9.4169H. doi:10.1038/s41467-018-06473-x. ISSN 2041-1723. PMC 6177445. PMID 30301893.
- ^ Lozupone, Catherine A.; Stombaugh, Jesse; Gonzalez, Antonio; Ackermann, Gail; Wendel, Doug; Vázquez-Baeza, Yoshiki; Jansson, Janet K.; Gordon, Jeffrey I.; Knight, Rob (October 2013). "Meta-analyses of studies of the human microbiota". Genome Research. 23 (10): 1704–1714. doi:10.1101/gr.151803.112. ISSN 1549-5469. PMC 3787266. PMID 23861384.
- ^ "Comprehensive profiling of human-to-human oral and intestinal microbial transmission". News-Medical.net. 20 January 2023. Archived from the original on 16 February 2023. Retrieved 16 February 2023.
- ^ Valles-Colomer, Mireia; Blanco-Míguez, Aitor; Manghi, Paolo; et al. (February 2023). "The person-to-person transmission landscape of the gut and oral microbiomes". Nature. 614 (7946): 125–135. Bibcode:2023Natur.614..125V. doi:10.1038/s41586-022-05620-1. ISSN 1476-4687. PMC 9892008. PMID 36653448.
- ^ a b Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. (November 2018). "US Immigration Westernizes the Human Gut Microbiome". Cell. 175 (4): 962–972.e10. doi:10.1016/j.cell.2018.10.029. PMC 6498444. PMID 30388453.
- ^ a b Kaplan RC, Wang Z, Usyk M, Sotres-Alvarez D, Daviglus ML, Schneiderman N, et al. (November 2019). "Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity". Genome Biology. 20 (1): 219. doi:10.1186/s13059-019-1831-z. PMC 6824043. PMID 31672155.
- ^ Moraïs, Sarah; Winkler, Sarah; Zorea, Alvah; Levin, Liron; Nagies, Falk S. P.; Kapust, Nils; Lamed, Eva; Artan-Furman, Avital; Bolam, David N.; Yadav, Madhav P.; Bayer, Edward A.; Martin, William F.; Mizrahi, Itzhak (15 March 2024). "Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans". Science. 383 (6688): eadj9223. Bibcode:2024Sci...383j9223M. doi:10.1126/science.adj9223. ISSN 0036-8075. PMC 7615765. PMID 38484069.
- ^ Stewart2024-03-14T18:00:00+00:00, Linda. "Cellulose-degrading gut bacteria found in the human gut, although at lower levels in industrialized countries". The Microbiologist. Retrieved 24 March 2024.
{{cite web}}: CS1 maint: numeric names: authors list (link)
External links
[edit]- The Secret World Inside You Exhibit 2015–2016, American Museum of Natural History
- FAQ: Human Microbiome, January 2014 American Society For Microbiology